3NA7
 
 | | 2.2 Angstrom Structure of the HP0958 Protein from Helicobacter pylori CCUG 17874 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HP0958, MAGNESIUM ION, ... | | Authors: | Caly, D.L, O'Toole, P.W, Moore, S.A. | | Deposit date: | 2010-06-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2-A Structure of the HP0958 Protein from Helicobacter pylori Reveals a Kinked Anti-Parallel Coiled-Coil Hairpin Domain and a Highly Conserved Zn-Ribbon Domain
J.Mol.Biol., 403, 2010
|
|
5H9D
 
 | | Crystal structure of Heptaprenyl Diphosphate Synthase from Staphylococcus aureus | | Descriptor: | C-terminal peptide from Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, Farnesyl pyrophosphate synthetase, Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, ... | | Authors: | Wei, H.L, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase
ChemMedChem, 11, 2016
|
|
7MFL
 
 | |
6CYU
 
 | | Crystal structure of CTX-M-14 S70G/N106S/D240G beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
5YL7
 
 | | Proteases from Pseudoalteromonas arctica PAMC 21717 (Pro21717) | | Descriptor: | CALCIUM ION, Copurified unknown peptide, Pseudoalteromonas arctica PAMC 21717 | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 angstrom resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme
PLoS ONE, 13, 2018
|
|
8OSE
 
 | | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), 1,2-ETHANEDIOL, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
7SQ9
 
 | | Cryo-EM structure of mouse temsirolimus/PI(3,5)P2-bound TRPML1 channel at 2.11 Angstrom resolution | | Descriptor: | (1R,2R,4S)-4-{(2R)-2-[(3S,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30S,34aS)-9,27-dihydroxy-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-1,5,11,28,29-pentaoxo-1,4,5,6,9,10,11,12,13,14,21,22,23,24,25,26,27,28,29,31,32,33,34,34a-tetracosahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontin-3-yl]propyl}-2-methoxycyclohexyl 3-hydroxy-2-(hydroxymethyl)-2-methylpropanoate, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MFK
 
 | |
6KF1
 
 | | Microbial Hormone-sensitive lipase- E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEXANOIC ACID, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
To Be Published
|
|
6GJ7
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 22 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[[1-(phenylmethyl)indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8EGV
 
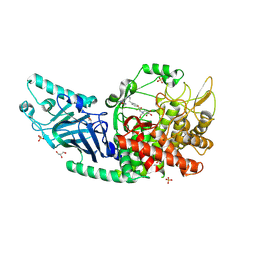 | | Co-crystal structure of Chaetomium glucosidase with compound 12 | | Descriptor: | (2R,3R,4R,5S)-1-{2-[4-(2-{[(5M)-3-chloro-5-(1,2,4-oxadiazol-3-yl)phenyl]amino}ethyl)phenyl]ethyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6NTN
 
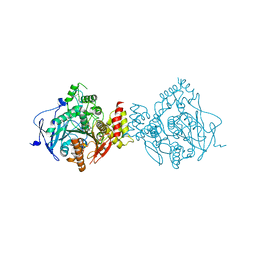 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-230 in Complex with the Reactivator, HI-6 | | Descriptor: | (S)-N-[(1E)-1-(diethylamino)ethylidene]-P-methylphosphonamidic fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
7YAW
 
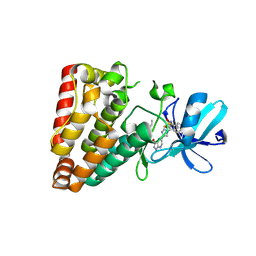 | | Crystal structure of ZAK in complex with compound YH-180 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[3-[[5-[1-[2,6-bis(fluoranyl)-3-[(3-phenylphenyl)sulfonylamino]phenyl]-1,2,3-triazol-4-yl]-1~{H}-pyrazolo[3,4-b]pyridin-3-yl]oxy]propyl]propanamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
5KJK
 
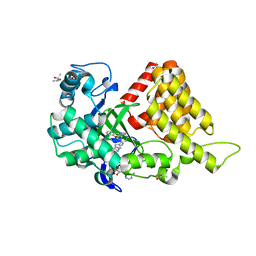 | | SMYD2 in complex with AZ370 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[2-[1-[2-(3,4-dichlorophenyl)ethyl]azetidin-3-yl]oxyphenyl]-~{N}-(3-pyrrolidin-1-ylpropyl)pyridine-4-carboxamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
8X3M
 
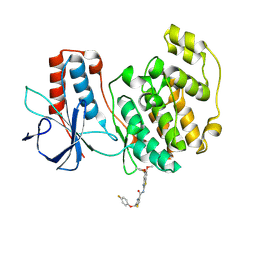 | | Crystal structure of p38alpha with an allosteric inhibitor 2 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-(5,6-dimethoxy-1,3-benzothiazol-2-yl)-2-[(4-fluoranylphenoxy)methyl]-1,3-thiazole-4-carboxamide | | Authors: | Hasegawa, S, Kinoshita, T. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct binding modes of a benzothiazole derivative confer structural bases for increasing ERK2 or p38 alpha MAPK selectivity.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
5HL9
 
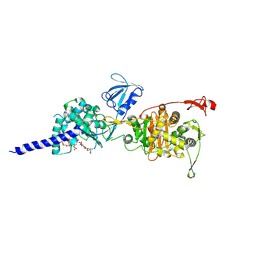 | | E. coli PBP1b in complex with acyl-ampicillin and moenomycin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into Inhibition of Escherichia coli Penicillin-binding Protein 1B.
J.Biol.Chem., 292, 2017
|
|
8ETL
 
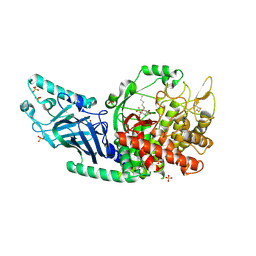 | | Co-crystal structure of Chaetomium glucosidase with compound 24 | | Descriptor: | (1S,2S,3R,4S,5S)-5-(butylamino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6T28
 
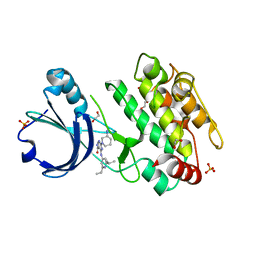 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
3MEM
 
 | |
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
9BDN
 
 | |
6K2H
 
 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
7M5N
 
 | | PCNA bound to peptide mimetic with linker | | Descriptor: | 1,3-dimethylbenzene, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | Authors: | Vandborg, B.A, Bruning, J.B. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
7Y99
 
 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
