8AKJ
 
 | | Acyl-enzyme complex of cephalothin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
4RHI
 
 | |
7TXS
 
 | | X-ray structure of the VioB N-aetyltransferase from Acinetobacter baumannii in the presence of a reaction intermediate | | Descriptor: | SODIUM ION, VioB, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (3R)-4-({3-[(2-{[(1S)-1-{[(2R,3S,4S,5R,6R)-4,5-dihydroxy-6-{[(R)-hydroxy{[(R)-hydroxy{[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)oxolan-2-yl]methoxy}phosphoryl]oxy}phosphoryl]oxy}-2-methyloxan-3-yl]amino}ethyl]sulfanyl}ethyl)amino]-3-oxopropyl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate (non-preferred name) | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
175L
 
 | |
5WI9
 
 | | Crystal structure of KL with an agonist Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 39F7 Fab heavy chain, ... | | Authors: | Johnstone, S, Min, X, Wang, Z. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Agonistic beta-Klotho antibody mimics fibroblast growth factor 21 (FGF21) functions.
J. Biol. Chem., 293, 2018
|
|
5F9S
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.7 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
6HSQ
 
 | |
6TPR
 
 | | PqsR (MvfR) bound to inhibitory compound 40 | | Descriptor: | 2-[(5-methyl-[1,2,4]triazino[5,6-b]indol-3-yl)sulfanyl]-~{N}-(4-pyridin-2-yloxyphenyl)ethanamide, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm GrownPseudomonas aeruginosa.
Front Chem, 8, 2020
|
|
8AY8
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB9-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
5FWS
 
 | | Wnt modulator Kremen crystal form I at 1.90A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
9BOL
 
 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | (4R)-1-[(2S)-2-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-3,3-dimethylbutanoyl]-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potency-Enhanced Peptidomimetic VHL Ligands with Improved Oral Bioavailability.
J.Med.Chem., 67, 2024
|
|
158L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
4ZEC
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocin 84 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
6E1H
 
 | | Structure of 2:1 human Ptch1-Shh-N complex | | Descriptor: | CALCIUM ION, PALMITIC ACID, Protein patched homolog 1, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex.
Science, 362, 2018
|
|
5ITD
 
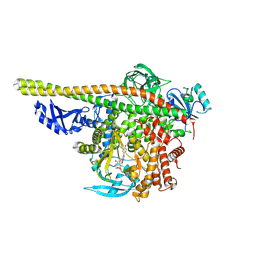 | | Crystal structure of PI3K alpha with PI3K delta inhibitor | | Descriptor: | 5-{4-[3-(4-acetylpiperazine-1-carbonyl)phenyl]quinazolin-6-yl}-2-methoxypyridine-3-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-09-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Discovery and Pharmacological Characterization of Novel Quinazoline-Based PI3K Delta-Selective Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
6BQA
 
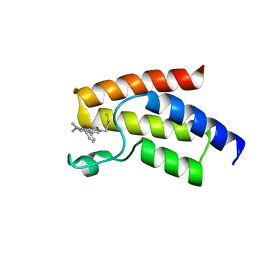 | | BRD9 bromodomain in complex with 3-(6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.031 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
8PDF
 
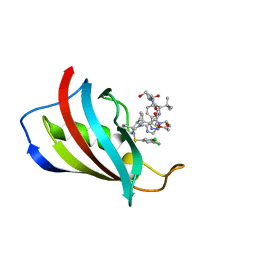 | | FKBP12 in complex with PROTAC 6a2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[4-[(1~{S})-1-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6TAA
 
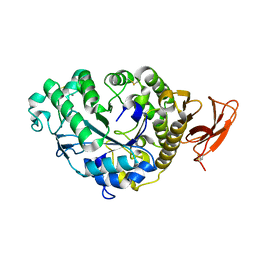 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF ASPERGILLUS ORYZAE (TAKA) ALPHA-AMYLASE: AN APPLICATION OF THE SIMULATED-ANNEALING METHOD | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Swift, H.J, Brady, L, Derewenda, Z.S, Dodson, E.J, Turkenburg, J.P, Wilkinson, A.J. | | Deposit date: | 1992-08-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and molecular model refinement of Aspergillus oryzae (TAKA) alpha-amylase: an application of the simulated-annealing method.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
4Z9N
 
 | |
9BRB
 
 | |
9GGY
 
 | | Human KRas4A (GDP) in complex with compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 12-(1-aza-5-azanidaspiro[4.5]decan-8-yl)-18-[(1-chloro-3-hydroxy-1,2,3-benzothiadiazol-5-yl)-lambda4-sulfanylidene]-4-hydroxy-3,3-dioxo-3lambda6-thia-1,4,12,17-tetraza-2lambda6-thia-6,10,19-triaza-3, GTPase KRas, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.266 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
5N0G
 
 | | Crystal Structure of CobH T85A (precorrin-8x methyl mutase) complexed with C5 allyl-HBA | | Descriptor: | 3-[(1~{R},2~{S},3~{S},4~{Z},7~{S},8~{S},9~{Z},15~{R},17~{R},18~{R},19~{R})-2,7,18-tris(2-hydroxy-2-oxoethyl)-3,13,17-tris(3-hydroxy-3-oxopropyl)-1,2,7,12,12,15,17-heptamethyl-5-prop-2-enyl-3,8,15,18,19,21-hexahydrocorrin-8-yl]propanoic acid, GLYCEROL, Precorrin-8X methylmutase | | Authors: | Nemoto-Smith, E.H, Lawrence, A.D, Brown, D.G, Warren, M.J. | | Deposit date: | 2017-02-02 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Novel cobalamin analogues and their application in the trafficking of cobalamin in bacteria, worms and plants
To Be Published
|
|
8RGH
 
 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
9DEW
 
 | | Crystal structure of NDM-1 complexed with compound 2 | | Descriptor: | Metallo-beta-lactamase type 2, ZINC ION, [(7-cyclopropyl-1-methyl-2-oxo-1,2-dihydroquinolin-4-yl)methyl]phosphonic acid | | Authors: | Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2024-08-29 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hit to Lead Optimization of Heteroaryl Phosphonates Reversible, Broad-Spectrum Inhibitors of Serine and Metallo Carbapenemases.
To Be Published
|
|
