8UHA
 
 | |
8UHD
 
 | | Structure of paused transcription complex Pol II-DSIF-NELF - post-translocated | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-08 | | Release date: | 2024-03-20 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8UHG
 
 | |
8UI0
 
 | |
8UIS
 
 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8QSZ
 
 | | Structure of s. pombe RNA polymerase II in complex with DSIF and Rat1/Rai1 | | Descriptor: | 5'-3' exoribonuclease 2, DNA none template (34-MER), DNA template (40-MER), ... | | Authors: | Carrique, L, Kus, K, Vasiljeva, L, Grimes, J.M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | DSIF factor Spt5 coordinates transcription, maturation and exoribonucleolysis of RNA polymerase II transcripts.
To Be Published
|
|
8UOQ
 
 | | Composite map of PIC_delta_TFIIK form2 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 52, 2024
|
|
8UOT
 
 | | Composite map of PICdeltaTFIIK form1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 52, 2024
|
|
8UPO
 
 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UPR
 
 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UQL
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UQP
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UQM
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8WWP
 
 | | PNPase mutant of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8URH
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 27 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URI
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 27 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8WX0
 
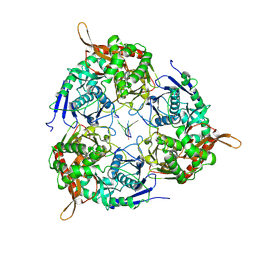 | | PNPase of M.tuberculosis with its RNA substrate | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase, RNA (24-mer) | | Authors: | Wang, N, Sheng, Y.N. | | Deposit date: | 2023-10-27 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8URY
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 30 nt long spacer, and fMet-tRNA in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URX
 
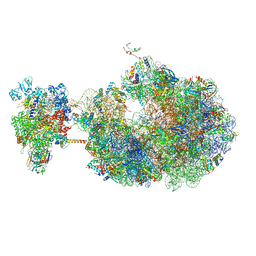 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 30 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8WXF
 
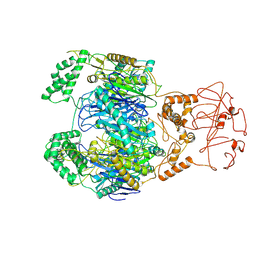 | | PNPase of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-29 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8QZZ
 
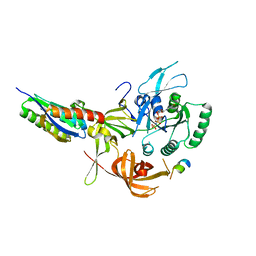 | | Crystal structure of human eIF2 alpha-gamma complexed with PPP1R15A_420-452 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Substrate recruitment via eIF2 gamma enhances catalytic efficiency of a holophosphatase that terminates the integrated stress response.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8R3V
 
 | |
8RAP
 
 | |
8RAM
 
 | |
8RBO
 
 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
