8W11
 
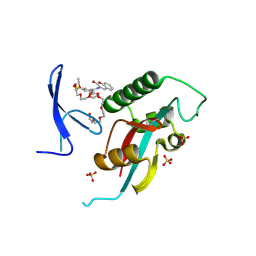 | | Structure of human PIN1 covalently derivatized with SuFEx compound | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-(fluorosulfonyl)-5-[(2-oxo-2H-1-benzopyran-3-yl)carbamoyl]benzoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Louie, G.V, Noel, J.P, Wang, W.-M, Kelly, J.W. | | Deposit date: | 2024-02-14 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human PIN1 covalently derivatized with SuFEx compound
To be published
|
|
2Y3P
 
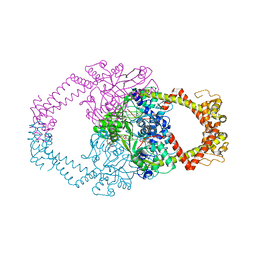 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
8VQI
 
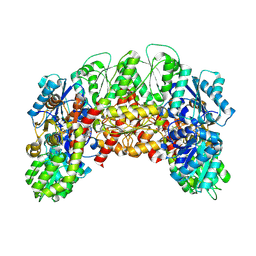 | |
5IJT
 
 | |
8VZ5
 
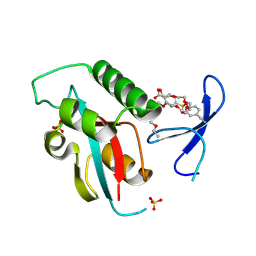 | | Structure of human PIN1 covalently derivatized with SuFEx compound | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-[fluoro(dihydroxy)-lambda~4~-sulfanyl]benzoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Louie, G.V, Noel, J.P, Wang, W.-M, Kelly, J.W. | | Deposit date: | 2024-02-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human PIN1 covalently derivatized with SuFEx compound
To be published
|
|
7ZET
 
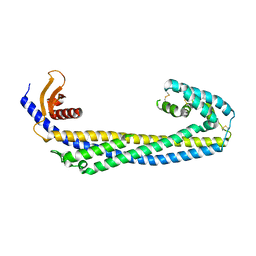 | | Crystal structure of human Clusterin, crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin, ... | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analyses define the molecular basis of clusterin chaperone function.
Nat.Struct.Mol.Biol., 2025
|
|
5IKG
 
 | |
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
8VUM
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
7ZEU
 
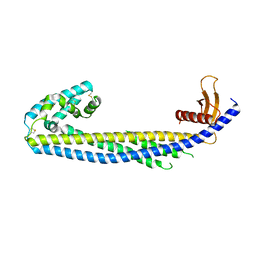 | | Crystal structure of human Clusterin, crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analyses define the molecular basis of clusterin chaperone function.
Nat.Struct.Mol.Biol., 2025
|
|
8VQH
 
 | |
5VZ3
 
 | |
6VPQ
 
 | |
8VIB
 
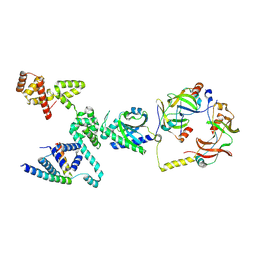 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
8VU9
 
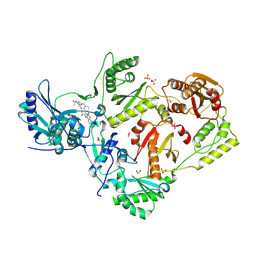 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
7MHL
 
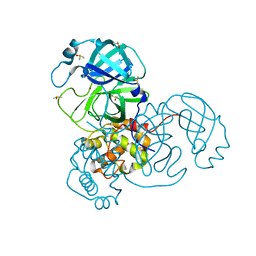 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
5IKX
 
 | |
7MQV
 
 | | Crystal structure of truncated (ACT domain removed) prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shabalin, I.G, Gritsunov, A, Gabryelska, A, Czub, M.P, Grabowski, M, Cooper, D.R, Christendat, D, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of Bacillus anthracis prephenate dehydrogenase identified an ACT regulatory domain and a novel mode of metabolic regulation for proteins within the prephenate dehydrogenase family of enzyme
to be published
|
|
8VUB
 
 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
5IL9
 
 | | Crystal structure of Deg9 | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
8VYS
 
 | | Cryo-EM Structure of the BRAF V600E monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
5ILM
 
 | | H64A sperm whale myoglobin with a Fe-chlorophenyl moiety | | Descriptor: | (4-chlorophenyl)[3,3'-(7,12-diethenyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)di(propanoato)(2-)]iron, GLYCEROL, Myoglobin, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
7MQ5
 
 | | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14 | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14
To Be Published
|
|
7MQN
 
 | |
