6NJA
 
 | | Structure of WT RET protein tyrosine kinase domain at 1.92A resolution. | | Descriptor: | ADENINE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6H09
 
 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2018-07-06 | | Release date: | 2018-08-15 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
7OTW
 
 | | DNA-PKcs in complex with AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
8S90
 
 | |
8S8Z
 
 | | Cryo-EM structure of octameric human CALHM1 (I109W) in complex with ruthenium red | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calcium homeostasis modulator protein 1, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | Authors: | Syrjanen, J.L, Furukawa, H. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structure of human CALHM1 reveals key locations for channel regulation and blockade by ruthenium red.
Nat Commun, 14, 2023
|
|
8KDA
 
 | | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, MAGNESIUM ION, RNA-free ribonuclease P | | Authors: | Teramoto, T, Adachi, N, Yokogawa, T, Koyasu, T, Mayanagi, K, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis of transfer RNA processing by bacterial minimal RNase P.
Nat Commun, 16, 2025
|
|
4WJ7
 
 | | CCM2 PTB domain in complex with KRIT1 NPxY/F3 | | Descriptor: | KRIT1 NPxY/F3, Malcavernin | | Authors: | Fisher, O.S, Liu, W, Zhang, R, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Structural Basis for the Disruption of the Cerebral Cavernous Malformations 2 (CCM2) Interaction with Krev Interaction Trapped 1 (KRIT1) by Disease-associated Mutations.
J.Biol.Chem., 290, 2015
|
|
8I6A
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Orotic acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, OROTIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
9GRG
 
 | | StmPr1, Stenotrophomonas maltophilia Protease 1, 36 kDa alkine serine protease in complex with PMSF | | Descriptor: | Alkaline serine protease, CALCIUM ION, GLYCEROL, ... | | Authors: | Sommer, M, Outzen, L, Negm, A, Windhorst, S, Weber, W, Betzel, C. | | Deposit date: | 2024-09-11 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep, 15, 2025
|
|
8G6C
 
 | | GTP Cyclohydrolase-IB with manganese | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase FolE2, MANGANESE (II) ION | | Authors: | McWhorter, K.L, Amaya Lopez, C.Y, Davis, K.M. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Combatting melioidosis with chemical synthetic lethality.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9G9U
 
 | | The structure of XynX, a NIF3 family protein from Geobacillus proteiniphilus T-6 | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase 1 type 2 homolog, ZINC ION | | Authors: | Hadad, N, Pomyalov, S, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2024-07-25 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of XynX, a NIF3 family protein from Geobacillus proteiniphilus T-6
To Be Published
|
|
8G6T
 
 | | Bromodomain of CBP liganded with inhibitor iCBP2 | | Descriptor: | (6S)-1-(3,4-dibromophenyl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein, NICKEL (II) ION | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8DOJ
 
 | | Dehaloperoxidase B in complex with 3,3'-Biphenol | | Descriptor: | (1P)-[1,1'-biphenyl]-3,3'-diol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
5IDY
 
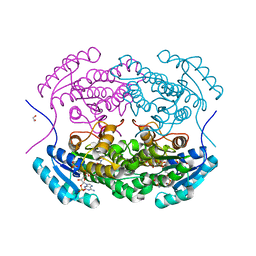 | |
6Z5B
 
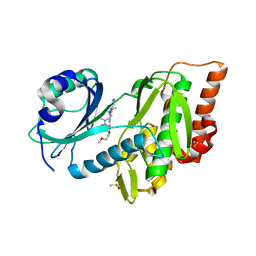 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003128 | | Descriptor: | 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003128
To Be Published
|
|
6NQW
 
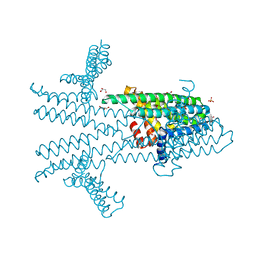 | | Flagellar protein FcpA from Leptospira biflexa - hexagonal form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Flagellar coiling protein A, GLYCEROL, ... | | Authors: | San Martin, F, Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
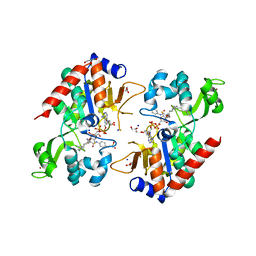
6NR0
 
 | |
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
6ZAB
 
 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (P6422 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Naretto, A, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6NQT
 
 | | GalNac-T2 soaked with UDP-sugar | | Descriptor: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{R})-3-(hex-5-ynoylamino)-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Fernandez, D, Bertozzi, C.R, Schumann, B, Agbay, A. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XPM
 
 | | Ancestral ADH WT | | Descriptor: | 1,2-ETHANEDIOL, A64 | | Authors: | Chen, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2022-05-04 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a versatile ancestral ADH with high activity and thermostability
To Be Published
|
|
7ZB0
 
 | | macrocyclase OphP with 15mer | | Descriptor: | 1,2-ETHANEDIOL, 15mer, BICARBONATE ION, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
6WYB
 
 | | RTX (Reverse Transcription Xenopolymerase) in complex with an RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA polymerase, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
