2M4H
 
 | | Solution structure of the Core Domain (10-76) of the Feline Calicivirus VPg protein | | Descriptor: | Feline Calicivirus VPg protein | | Authors: | Kwok, R.N, Leen, E.N, Birtley, J.R, Prater, S.N, Simpson, P.J, Curry, S, Matthews, S, Marchant, J. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
2M4I
 
 | |
2M4J
 
 | | 40-residue beta-amyloid fibril derived from Alzheimer's disease brain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Lu, J, Qiang, W, Meredith, S.C, Yau, W, Schweiters, C.D, Tycko, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of beta-Amyloid Fibrils in Alzheimer's Disease Brain Tissue.
Cell(Cambridge,Mass.), 154, 2013
|
|
2M4K
 
 | | Solution structure of the delta subunit of RNA polymerase from Bacillus subtilis | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Papouskova, V, Novacek, J, Kaderavek, P, Zidek, L, Rabatinova, A, Sanderova, H, Krasny, L, Sklenar, V. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Study of the Partially Disordered Full-Length delta Subunit of RNA Polymerase from Bacillus subtilis.
Chembiochem, 14, 2013
|
|
2M4L
 
 | |
2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2M4N
 
 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
2M4P
 
 | |
2M4Q
 
 | | NMR structure of E. coli ribosomela decoding site with apramycin | | Descriptor: | APRAMYCIN, RNA (27-MER) | | Authors: | Puglisi, J.D, Tsai, A, Marshall, R, Viani, E. | | Deposit date: | 2013-02-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The impact of aminoglycosides on the dynamics of translation elongation.
Cell Rep, 3, 2013
|
|
2M4V
 
 | | Mycobacterium tuberculosis RNA polymerase binding protein A (RbpA) and its interactions with sigma factors | | Descriptor: | Putative uncharacterized protein | | Authors: | Bortoluzzi, A, Muskett, F.W, Waters, L.C, Addis, P.W, Rieck, B, Munder, T, Schleier, S, Forti, F, Ghisotti, D, Carr, M.D, O'Hare, H.M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors.
J.Biol.Chem., 288, 2013
|
|
2M4W
 
 | |
2M4X
 
 | |
2M4Y
 
 | |
2M4Z
 
 | |
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | Descriptor: | Mu-theraphotoxin-Hh2a | | Authors: | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
2M51
 
 | |
2M52
 
 | |
2M53
 
 | |
2M54
 
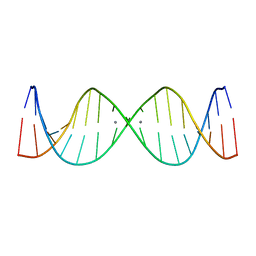 | | Refined NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Kumbhar, S, Johannsen, S, Sigel, R.K, Waller, M.P, Mueller, J. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A QM/MM refinement of an experimental DNA structure with metal-mediated base pairs.
J.Inorg.Biochem., 127, 2013
|
|
2M55
 
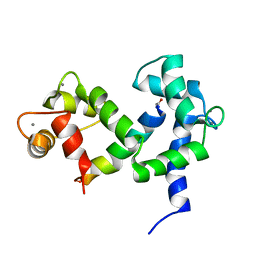 | | NMR structure of the complex of an N-terminally acetylated alpha-synuclein peptide with calmodulin | | Descriptor: | Alpha-synuclein, CALCIUM ION, Calmodulin | | Authors: | Gruschus, J.M, Yap, T, Pistolesi, S, Maltsev, A.S, Lee, J.C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Calmodulin Complexed to an N-Terminally Acetylated alpha-Synuclein Peptide.
Biochemistry, 52, 2013
|
|
2M56
 
 | | The structure of the complex of cytochrome P450cam and its electron donor putidaredoxin determined by paramagnetic NMR spectroscopy | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hiruma, Y, Hass, M.A.S, Ubbink, M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
2M57
 
 | |
2M58
 
 | | Structure of 2'-5' AG1 lariat forming ribozyme in its inactive state | | Descriptor: | RNA (59-MER) | | Authors: | Carlomagno, T, Amata, I, Codutti, L, Falb, M, Fohrer, J, Simon, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural principles of RNA catalysis in a 2'-5' lariat-forming ribozyme.
J.Am.Chem.Soc., 135, 2013
|
|
2M59
 
 | |
2M5A
 
 | | Protein A binding by an engineered Affibody molecule | | Descriptor: | Immunoglobulin G-binding protein A, ZpA963 | | Authors: | Hard, T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-affinity binding to staphylococcal protein A by an engineered dimeric Affibody molecule.
Protein Eng.Des.Sel., 26, 2013
|
|
