5JY5
 
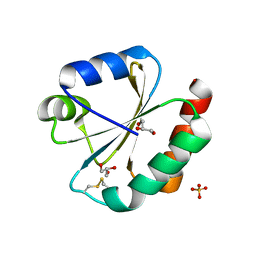 | | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Bravo-Chaucanes, C.P, Abadio, A.K.R, Kioshima, E.S, Felipe, M.S.S, Barbosa, J.A.R.G. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution
To Be Published
|
|
7THS
 
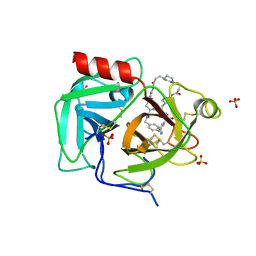 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2S)-butane-1,2-diol, (6S,9R,20R,23S)-N-{[4-(aminomethyl)phenyl]methyl}-20-[(benzenesulfonyl)amino]-3,13,21-trioxo-2,6,9,14,22-pentaazatetracyclo[23.2.2.2~6,9~.2~15,18~]tritriaconta-1(27),15,17,25,28,30-hexaene-23-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
6AEX
 
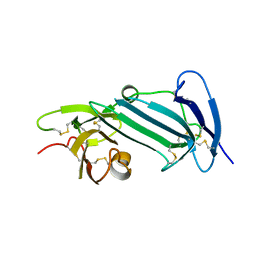 | | Crystal structure of unoccupied murine uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-08-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of the unoccupied murine urokinase-type plasminogen activator receptor (uPAR) reveals a tightly packed DII-DIII unit.
Febs Lett., 593, 2019
|
|
8DYC
 
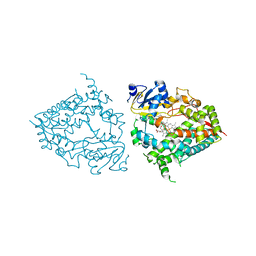 | | Human CYP3A4 bound to a substrate | | Descriptor: | 2-butyl-6-(butylamino)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-08-04 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CYP3A4 Complexed with Fluorol Identifies the Substrate Access Channel as a High-Affinity Ligand Binding Site.
Int J Mol Sci, 23, 2022
|
|
5JYL
 
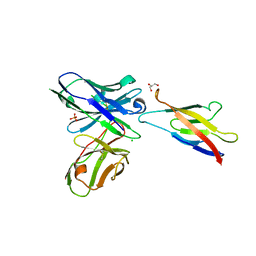 | | Human P-cadherin MEC1 with scFv TSP7 bound | | Descriptor: | CHLORIDE ION, Cadherin-3, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
8IZC
 
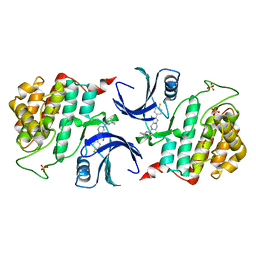 | | Human CK1 Delta Kinase structure bound to Inhibitor | | Descriptor: | Casein kinase I isoform delta, SULFATE ION, ~{N}5-~{tert}-butyl-2-(3-chloranyl-4-fluoranyl-phenyl)-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-3,5-dicarboxamide | | Authors: | Ghosh, K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Discovery of BMS-986164, a Potent, Selective and Orally Efficacious CK1d/e/a Inhibitor from Pyrazolo-Piperazine Chemotype
To Be Published
|
|
5KCX
 
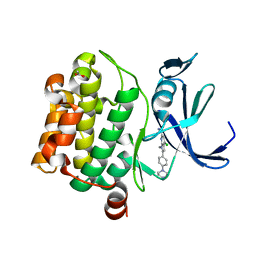 | | Pim-1 kinase in Complex with a Selective N-substituted 7-azaindole Inhibitor | | Descriptor: | 4-chloranyl-1-methyl-2-[4-(4-methylpiperazin-1-yl)phenyl]pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Mechin, I, McLean, L.R, Zhang, Y, Wang, R. | | Deposit date: | 2016-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8DOH
 
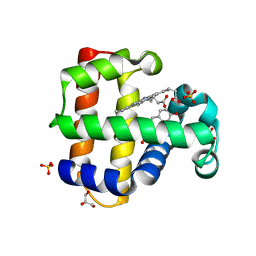 | | Dehaloperoxidase B in complex with Bisphenol F | | Descriptor: | 4,4'-methylenediphenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
5KE8
 
 | | mouse Klf4 E446P ZnF1-3 and MpG/MpG sequence DNA complex structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), Krueppel-like factor 4, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
5KER
 
 | | Deer mouse recombinant hemoglobin from high altitude species | | Descriptor: | Alpha-globin, Beta globin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Inoguchi, N, Natarajan, C, Storz, J.F, Moriyama, H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Alteration of the alpha 1 beta 2/ alpha 2 beta 1 subunit interface contributes to the increased hemoglobin-oxygen affinity of high-altitude deer mice.
PLoS ONE, 12, 2017
|
|
6KUD
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 3) | | Descriptor: | GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
8JG8
 
 | |
8DOI
 
 | | Dehaloperoxidase B in complex with 2,2'-Biphenol | | Descriptor: | 2-(2-hydroxyphenyl)phenol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
8DOG
 
 | | Dehaloperoxidase B in complex with Bisphenol E | | Descriptor: | 4-[1-(4-hydroxyphenyl)ethyl]phenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
5KHI
 
 | | HCN2 CNBD in complex with purine riboside-3', 5'-cyclic monophosphate (cPuMP) | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Purine riboside-3',5'-cyclic monophosphate | | Authors: | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | Deposit date: | 2016-06-14 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
8DOJ
 
 | | Dehaloperoxidase B in complex with 3,3'-Biphenol | | Descriptor: | (1P)-[1,1'-biphenyl]-3,3'-diol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
6A4O
 
 | | HEWL crystals soaked in 2.5M GuHCl for 20 minutes | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANIDINE, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
6KQU
 
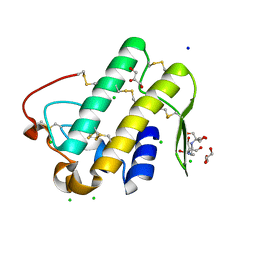 | | Crystal structure of phospholipase A2 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, T, Liu, J. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residue Asn21 acts as a switch for calcium binding to modulate the enzymatic activity of human phospholipase A2 group IIE.
Biochimie, 176, 2020
|
|
6KUB
 
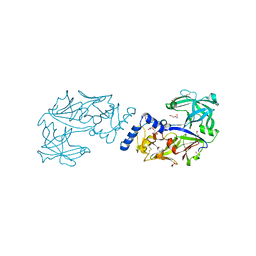 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
7U08
 
 | |
5K5N
 
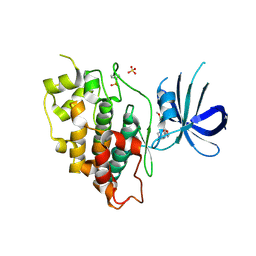 | | Crystal structure of GSK-3beta complexed with PF-04802367, a highly selective brain-penetrant kinase inhibitor | | Descriptor: | 5-(3-chloranyl-4-methoxy-phenyl)-~{N}-[3-(1,2,4-triazol-1-yl)propyl]-1,3-oxazole-4-carboxamide, Glycogen synthase kinase-3 beta, SULFATE ION | | Authors: | Kurumbail, R.G, Chang, J.S. | | Deposit date: | 2016-05-23 | | Release date: | 2016-09-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Highly Selective Glycogen Synthase Kinase-3 Inhibitor (PF-04802367) That Modulates Tau Phosphorylation in the Brain: Translation for PET Neuroimaging.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5KAO
 
 | | Crystal structure of wild type HIV-1 protease in complex with GRL-10413 | | Descriptor: | [(3~{a}~{S},4~{R},6~{a}~{R})-2,3,3~{a},4,5,6~{a}-hexahydrofuro[2,3-b]furan-4-yl] ~{N}-[(2~{S},3~{R})-1-(3-chloranyl-4-methoxy-phenyl)-4-[(4-methoxyphenyl)sulfonyl-(2-methylpropyl)amino]-3-oxidanyl-butan-2-yl]carbamate, protease | | Authors: | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-06-01 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.
Antimicrob.Agents Chemother., 60, 2016
|
|
7TZM
 
 | |
7U00
 
 | |
8DL4
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH Courmarin-containing INHIBITOR | | Descriptor: | 7-(diethylamino)-N-[(2S)-2-(2,4-difluorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]-2-oxo-2H-1-benzopyran-3-carboxamide, Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, ... | | Authors: | Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | S. CEREVISIAE CYP51 COMPLEXED WITH Courmarin-containing INHIBITOR
To Be Published
|
|
