2K8I
 
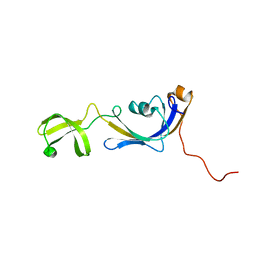 | | Solution structure of E.Coli SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Weininger, U, Balbach, J. | | Deposit date: | 2008-09-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of SlyD from Escherichia coli: spatial separation of prolyl isomerase and chaperone function.
J.Mol.Biol., 387, 2009
|
|
2IF4
 
 | |
2FKE
 
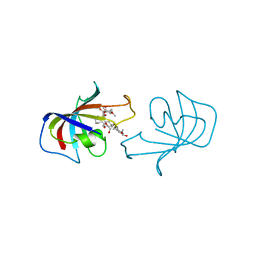 | | FK-506-BINDING PROTEIN: THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX WITH THE ANTAGONIST L-685,818 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Becker, J.W, Mckeever, B.M, Rotonda, J. | | Deposit date: | 1993-01-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818.
J.Biol.Chem., 268, 1993
|
|
2FAP
 
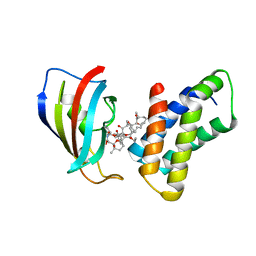 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-(C16)-ETHOXY RAPAMYCIN COMPLEX INTERACTING WITH HUMA | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FRAP | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2F4E
 
 | |
2F2D
 
 | | Solution structure of the FK506-binding domain of human FKBP38 | | Descriptor: | 38 kDa FK-506 binding protein homolog, FKBP38 | | Authors: | Maestre-Martinez, M, Edlich, F, Jarczowski, F, Weiwad, M, Fischer, G, Luecke, C. | | Deposit date: | 2005-11-16 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the FK506-Binding Domain of Human FKBP38
J.BIOMOL.NMR, 34, 2006
|
|
2DG9
 
 | |
2DG4
 
 | | FK506-binding protein mutant WF59 complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2DG3
 
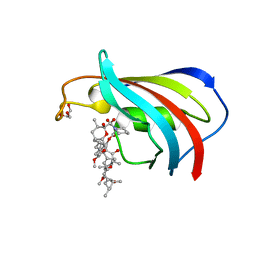 | | Wildtype FK506-binding protein complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2D9F
 
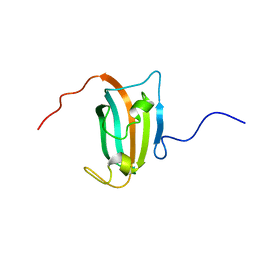 | | Solution structure of RUH-047, an FKBP domain from human cDNA | | Descriptor: | FK506-binding protein 8 variant | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-06-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RUH-047, an FKBP domain from human cDNA
To be Published
|
|
2AWG
 
 | | Structure of the PPIase domain of the Human FK506-binding protein 8 | | Descriptor: | 38 kDa FK-506 binding protein | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Finerty, P, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human FK-506 binding protein 8
To be Published
|
|
1YAT
 
 | |
1Y0O
 
 | |
1W26
 
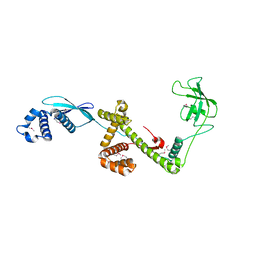 | | Trigger Factor in Complex with the Ribosome forms a Molecular Cradle for Nascent Proteins | | Descriptor: | TRIGGER FACTOR | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-06-28 | | Release date: | 2004-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
1U79
 
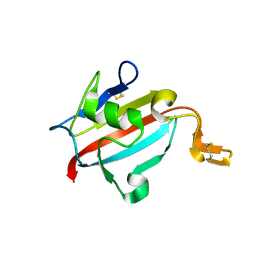 | | Crystal structure of AtFKBP13 | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase 3 | | Authors: | Gopalan, G, Swaminathan, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TCO
 
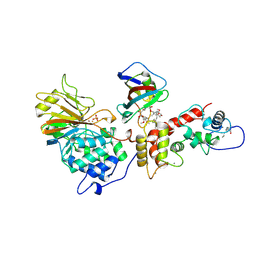 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
1T11
 
 | | Trigger Factor | | Descriptor: | Trigger factor | | Authors: | Ludlam, A.V, Moore, B.A, Xu, Z. | | Deposit date: | 2004-04-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1ROU
 
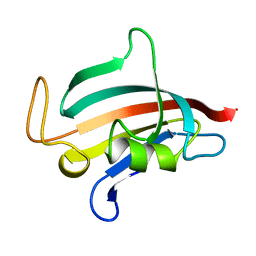 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, 22 STRUCTURES | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
1ROT
 
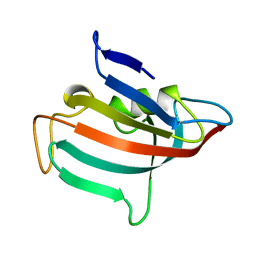 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
1R9H
 
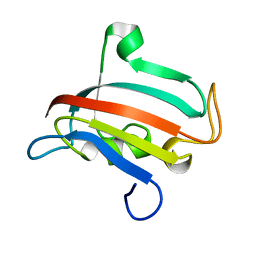 | |
1QZ2
 
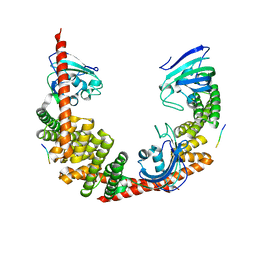 | | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | | Descriptor: | 5-mer peptide from Heat shock protein HSP 90, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Liu, Y, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-09-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1QPL
 
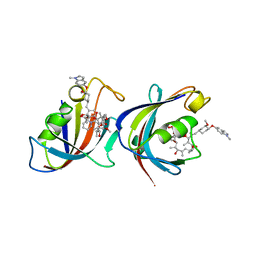 | | FK506 BINDING PROTEIN (12 KDA, HUMAN) COMPLEX WITH L-707,587 | | Descriptor: | C32-O-(1-METHYL-INDOL-5-YL) 18-HYDROXY-ASCOMYCIN, PROTEIN (FK506-BINDING PROTEIN) | | Authors: | Becker, J.W, Rotonda, J. | | Deposit date: | 1999-05-25 | | Release date: | 1999-08-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein.
J.Med.Chem., 42, 1999
|
|
1QPF
 
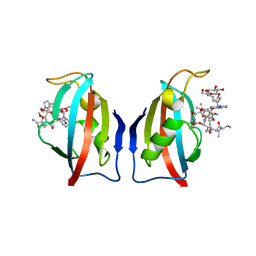 | | FK506 BINDING PROTEIN (12 KDA, HUMAN) COMPLEX WITH L-709,858 | | Descriptor: | C32-O-(1-ETHYL-INDOL-5-YL)ASCOMYCIN, PROTEIN (FK506-BINDING PROTEIN), heptyl beta-D-glucopyranoside | | Authors: | Becker, J.W, Rotonda, J. | | Deposit date: | 1999-05-24 | | Release date: | 1999-08-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein.
J.Med.Chem., 42, 1999
|
|
1Q6U
 
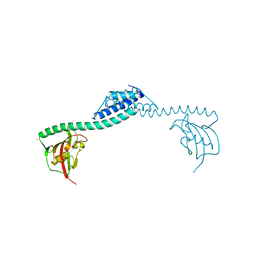 | | Crystal structure of FkpA from Escherichia coli | | Descriptor: | CESIUM ION, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-14 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6I
 
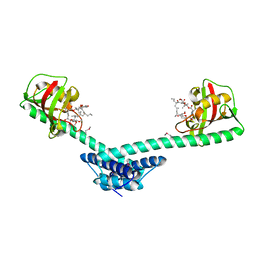 | | Crystal structure of a truncated form of FkpA from Escherichia coli, in complex with immunosuppressant FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
