3RGF
 
 | | Crystal Structure of human CDK8/CycC | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2011-04-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder.
J.Mol.Biol., 412, 2011
|
|
6KTO
 
 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
2NPQ
 
 | |
2IAJ
 
 | |
4Q21
 
 | |
4XAH
 
 | |
9II2
 
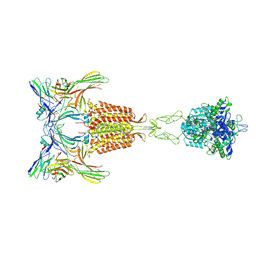 | | Cryo-EM Structure of the 2:2 Complex of mGlu3 and beta-arrestin1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-arrestin-1, CHOLESTEROL, ... | | Authors: | Wen, T.L, Du, M, Yang, X, Shen, Y.Q. | | Deposit date: | 2024-06-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Nat.Chem.Biol., 2025
|
|
9II3
 
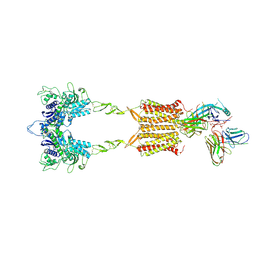 | | Cryo-EM Structure of the 2:1 Complex of mGlu3 and beta-arrestin1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-arrestin-1, CHOLESTEROL, ... | | Authors: | Wen, T.L, Du, M, Yang, X, Shen, Y.Q. | | Deposit date: | 2024-06-19 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Nat.Chem.Biol., 2025
|
|
4DTR
 
 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTJ
 
 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite an Abasic Site and ddT/dA as the Penultimate Base-pair | | Descriptor: | CALCIUM ION, DNA polymerase, DNA primer, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTO
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddA/dT as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
6SKL
 
 | | Cryo-EM structure of the CMG Fork Protection Complex at a replication fork - Conformation 1 | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA fork, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
6ZWR
 
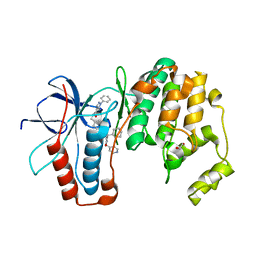 | | p38a bound with SR92 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-(pyridin-4-ylmethylamino)butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
3BRD
 
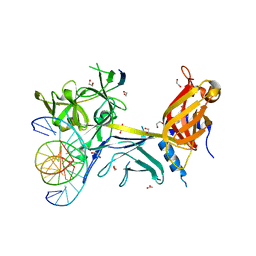 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
7R1Y
 
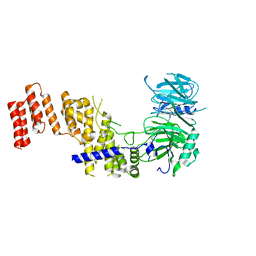 | |
6BCB
 
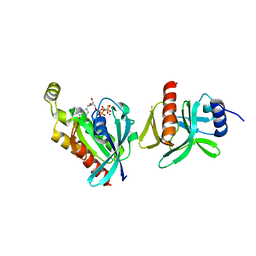 | |
4DTN
 
 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite an Abasic Site and ddA/dT as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTU
 
 | | RB69 DNA Polymerase Ternary Complex with dGTP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
5MHC
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, LYS-LEU-MET-PHE-LYS-TPO-GLU-GLY-PRO-ASP-SER-ASP, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-11-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
4DTM
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddG/dC as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTP
 
 | | RB69 DNA Polymerase Ternary Complex with dGTP Opposite an Abasic Site and ddA/dT as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTS
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
5JHH
 
 | | Crystal structure of the ternary complex between the human RhoA, its inhibitor and the DH/PH domain of human ARHGEF11 | | Descriptor: | 3-{3-[ethyl(quinolin-2-yl)amino]phenyl}propanoic acid, GLYCEROL, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Lv, Z, Wang, R, Ma, L, Miao, Q, Wu, J, Yan, Z, Li, J, Miao, L, Wang, F. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and PDZRhoGEF
To Be Published
|
|
5MOC
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
6BCA
 
 | |
