3TM0
 
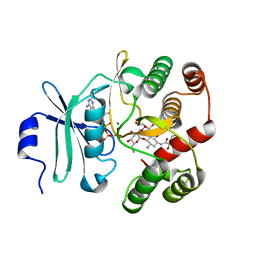 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Butirosin A Complex | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,6-dideoxy-alpha-D-glucopyranosyl)oxy]-2-hydroxy-3-(beta-D-xylofuranosyloxy)cyclohexyl]-2-hydroxybutanamide, Aminoglycoside 3'-phosphotransferase, MAGNESIUM ION, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2011-08-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of APH(3')-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics.
Antimicrob.Agents Chemother., 53, 2009
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
4Q5H
 
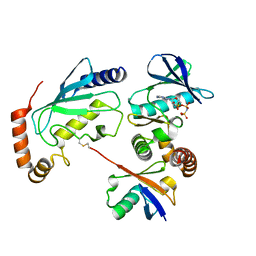 | |
4JXC
 
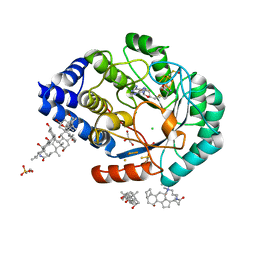 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | Descriptor: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6H79
 
 | |
3BDN
 
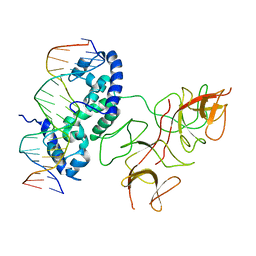 | | Crystal Structure of the Lambda Repressor | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DAP*DCP*DCP*DAP*DCP*DTP*DGP*DGP*DCP*DGP*DGP*DTP*DGP*DAP*DTP*DAP*DT)-3'), DNA (5'-D(*DTP*DAP*DTP*DAP*DTP*DCP*DAP*DCP*DCP*DGP*DCP*DCP*DAP*DGP*DTP*DGP*DGP*DTP*DAP*DT)-3'), Lambda Repressor | | Authors: | Stayrook, S.E, Jaru-Ampornpan, P, Hochschild, A, Lewis, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.909 Å) | | Cite: | Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding
Nature, 452, 2008
|
|
4J5J
 
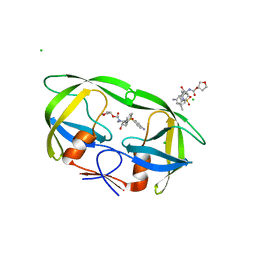 | |
3VH9
 
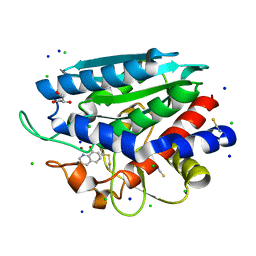 | | Crystal structure of Aeromonas proteolytica aminopeptidase complexed with 8-quinolinol | | Descriptor: | Bacterial leucyl aminopeptidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Hanaya, K, Suetsugu, M, Kobayashi, K, Yamato, I, Aoki, S. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Potent inhibition of dinuclear zinc(II) peptidase, an aminopeptidase from Aeromonas proteolytica, by 8-quinolinol derivatives: inhibitor design based on Zn(2+) fluorophores, kinetic, and X-ray crystallographic study.
J.Biol.Inorg.Chem., 17, 2012
|
|
2XWD
 
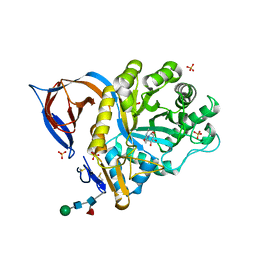 | | X-RAY STRUCTURE OF ACID-BETA-GLUCOSIDASE WITH 5N,6O-(N'-(N-OCTYL)IMINO)NOJIRIMYCIN IN THE ACTIVE SITE | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, GLUCOSYLCERAMIDASE, SULFATE ION, ... | | Authors: | Brumshtein, B, Aguilar-Moncayo, M, Benito, J.M, Ortiz Mellet, C, Garcia Fernandez, J.M, Silman, I, Shaaltiel, Y, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Cyclodextrin-Mediated Crystallization of Acid Beta-Glucosidase in Complex with Amphiphilic Bicyclic Nojirimycin Analogues.
Org.Biomol.Chem., 9, 2011
|
|
3E3K
 
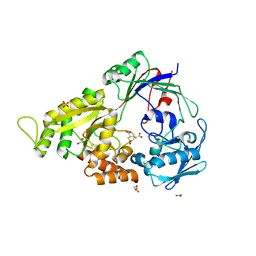 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (butane-1,2,4-tricarboxylate without nickel form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-08-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
4KII
 
 | | Beta-lactoglobulin in complex with Cp*Rh(III)Cl N,N-di(pyridin-2-yl)dodecanamide | | Descriptor: | Beta-lactoglobulin, [N,N-di(pyridin-2-yl-kappaN)dodecanamide]rhodium | | Authors: | Cherrier, M.V, Amara, P, Engilberge, S, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-02 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Enantioselectivity in the Transfer Hydrogenation of a Ketone Catalyzed by an Artificial Metalloenzyme
Eur.J.Inorg.Chem., 21, 2013
|
|
1DDI
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
5IPG
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure FFT-butyl (Hyperoxodized by t-butyl hydroperoxide) | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
1DDG
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
5IMD
 
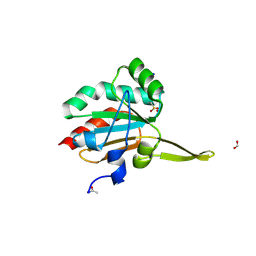 | | Xanthomonas campestris Peroxiredoxin Q - Structure F4 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, OXYGEN ATOM, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
1ETH
 
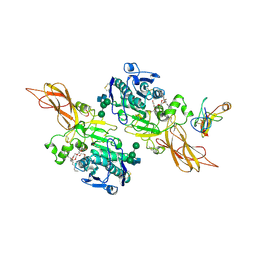 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
5INY
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F8 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
7UH9
 
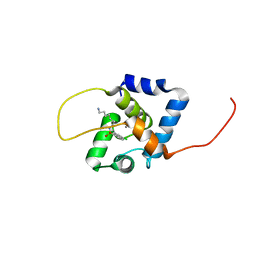 | | NMR structure of the cNTnC-cTnI chimera bound to W8 | | Descriptor: | CALCIUM ION, N-(7-aminoheptyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
7UHA
 
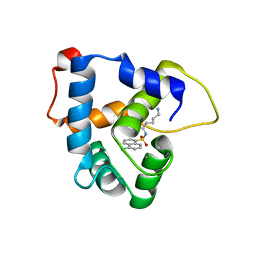 | | NMR structure of the cNTnC-cTnI chimera bound to W6 | | Descriptor: | CALCIUM ION, N-(5-aminopentyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
5IOX
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure LUss | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMV
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F6 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
2FQ5
 
 | |
5IM9
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F1 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F7 | | Descriptor: | Bacterioferritin comigratory protein, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
3DP8
 
 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (nickel butane-1,2,4-tricarboxylate form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
