7WNO
 
 | |
4Y6I
 
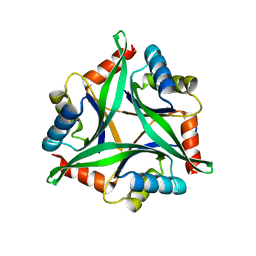 | |
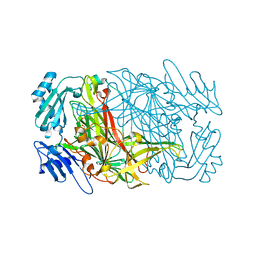
5WRE
 
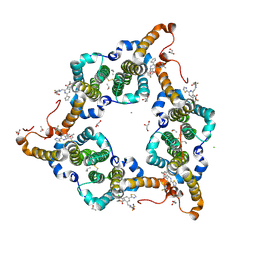 | | Hepatitis B virus core protein Y132A mutant in complex with heteroaryldihydropyrimidine (HAP_R01) | | Descriptor: | (2S)-1-[[(4R)-4-(2-chloranyl-4-fluoranyl-phenyl)-5-methoxycarbonyl-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-4,4-bis(fluoranyl)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-12-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
4YF5
 
 | |
4YKG
 
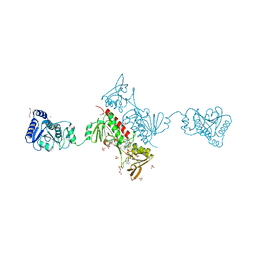 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NAD+ from Escherichia coli | | Descriptor: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
7Y5I
 
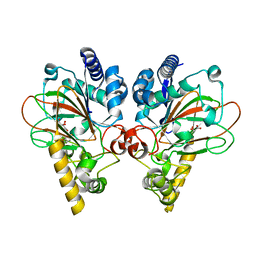 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | ARGININE, CmnC, FE (III) ION, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5P
 
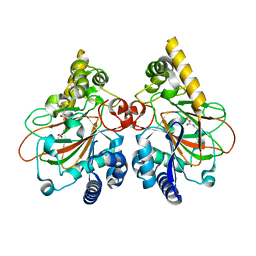 | | Crystal structure of CmnC in complex with L-arginine and alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, CmnC, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
5TQV
 
 | |
4YT4
 
 | |
4YB0
 
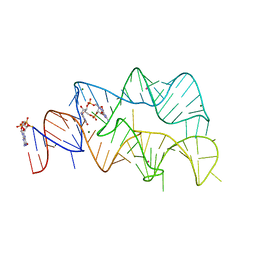 | | 3',3'-cGAMP riboswitch bound with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
5U1T
 
 | |
7MXE
 
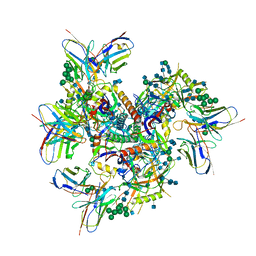 | | Ab1245 Fab in complex with BG505 SOSIP.664 and 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 G52K5 Fab heavy chain, ... | | Authors: | Abernathy, M.E, Bjorkman, P.J. | | Deposit date: | 2021-05-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody elicited by HIV-1 immunogen vaccination in macaques displaces Env fusion peptide and destroys a neutralizing epitope.
Npj Vaccines, 6, 2021
|
|
5U9K
 
 | |
5AFW
 
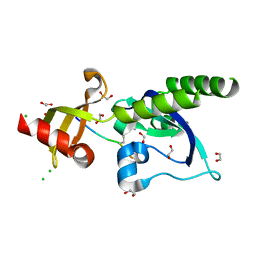 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | Authors: | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5A40
 
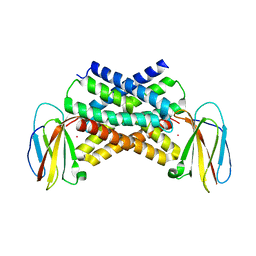 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | MERCURY (II) ION, MONOBODIES, PUTATIVE FLUORIDE ION TRANSPORTER CRCB | | Authors: | Stockbridge, R.B, Kolmakova-Partensky, L, Shane, T, Koide, A, Koide, S, Miller, C, Newstead, S. | | Deposit date: | 2015-06-04 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Nature, 525, 2015
|
|
5TPA
 
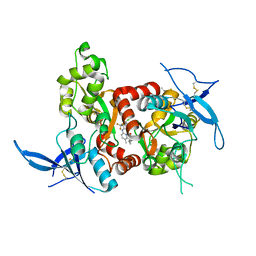 | | Structure of the human GluN1/GluN2A LBD in complex with compound 9 (GNE3500) | | Descriptor: | (1R,2R)-2-(2-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-7-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-6-yl)cyclopropane-1-carbonitrile, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
5AHT
 
 | |
5AIK
 
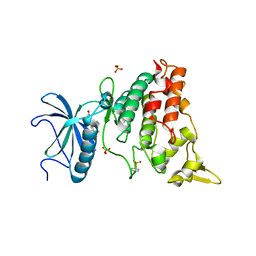 | | Human DYRK1A in complex with LDN-211898 | | Descriptor: | 4-(7-METHOXY-1-(TRIFLUOROMETHYL)-9H-PYRIDO[3,4-B]INDOL-9-yl)butan-1-amine, DYRK1A DUAL-SPECIFICITY TYROSINE-PHOSPHORYLATION REGULATED KINASE 1A, PHOSPHATE ION | | Authors: | Elkins, J.M, Soundararajan, M, Muniz, J.R.C, Cuny, G, Higgins, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dyrk1A with Ldn-211898
To be published
|
|
7XZ2
 
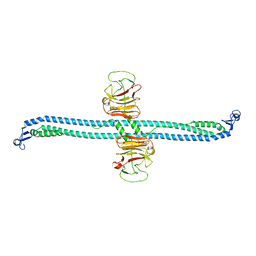 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7N1Z
 
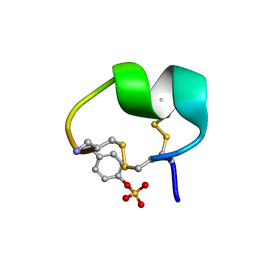 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7Y1Y
 
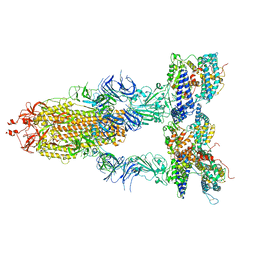 | | S-ECD (Omicron BA.2) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7N24
 
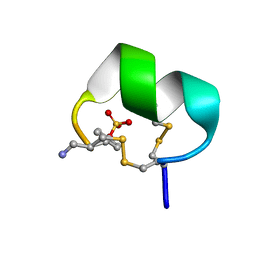 | | NMR structure of native EpI | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N25
 
 | | NMR structure of EpI-OH | | Descriptor: | Alpha-conotoxin EpI-OH | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N26
 
 | | NMR structure of EpI-[Y(SO3)15Y]-NH2 | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
5W6A
 
 | | HLA-C*06:02 presenting ARTELYRSL | | Descriptor: | ALA-ARG-THR-GLU-LEU-TYR-ARG-SER-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
