3EMQ
 
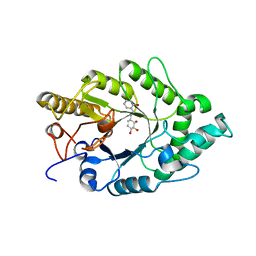 | | Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor | | Descriptor: | (1S,2S,3R,6R)-6-[(2-hydroxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
6SYS
 
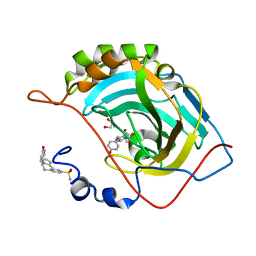 | | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2 | | Descriptor: | (4~{S})-4-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, GLYCEROL, ZINC ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2
To Be Published
|
|
3EAD
 
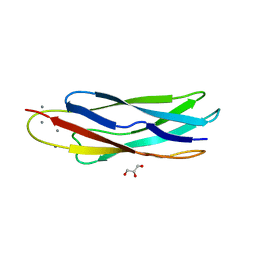 | | Crystal structure of CALX-CBD1 | | Descriptor: | CALCIUM ION, GLYCEROL, Na/Ca exchange protein | | Authors: | Zheng, L, Wang, M. | | Deposit date: | 2008-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
6SWQ
 
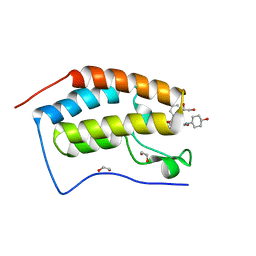 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
2P3H
 
 | | Crystal structure of the CorC_HlyC domain of a putative Corynebacterium glutamicum hemolysin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Uncharacterized CBS domain-containing protein | | Authors: | Cuff, M.E, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the CorC_HlyC domain of a putative Corynebacterium glutamicum hemolysin.
TO BE PUBLISHED
|
|
1MMU
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-glucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-D-glucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
3N3Z
 
 | | Crystal structure of PDE9A (E406A) mutant in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CHLORIDE ION, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Hou, J, Luo, H.-B, Chen, Y, Xu, J, Zhao, R, Zou, L. | | Deposit date: | 2010-05-21 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PDE9A (E406A) mutation in complex with IBMX
To be Published
|
|
6SXF
 
 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with Tamoxifen (2.8 Angstrom resolution) | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
3VPQ
 
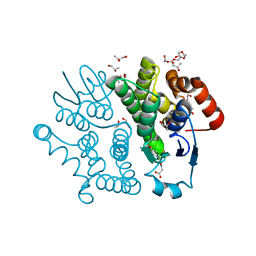 | | Crystal structure of Bombyx mori sigma-class glutathione transferase in complex with glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, Glutathione S-transferase sigma, ... | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A, Suzuki, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of a Bombyx mori sigma-class glutathione transferase exhibiting prostaglandin E synthase activity
Biochim.Biophys.Acta, 1830, 2013
|
|
3EDZ
 
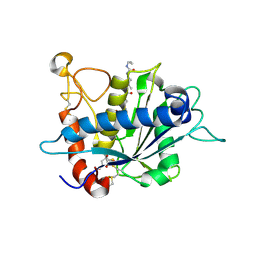 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | Descriptor: | ADAM 17, CITRIC ACID, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Mazzola, R.D, Zhu, Z, Sinning, L, McKittrick, B, Lavey, B, Spitler, J, Kozlowski, J, Neng-Yang, S, Zhou, G, Guo, Z, Orth, P, Madison, V, Sun, J, Lundell, D, Niu, X. | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3N2L
 
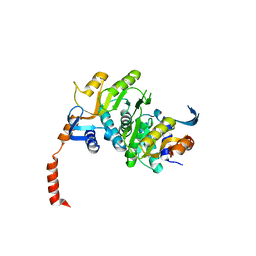 | | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase (pyrE) from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase
(pyrE) from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3E3U
 
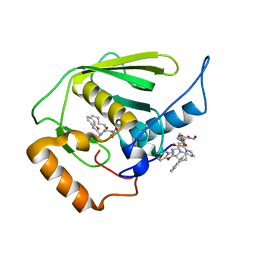 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1MQP
 
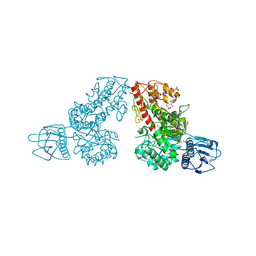 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
6DHM
 
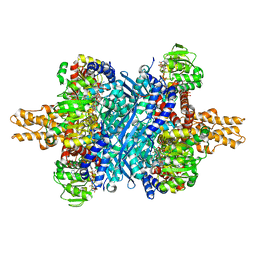 | | Bovine glutamate dehydrogenase complexed with zinc | | Descriptor: | GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis.
FEBS J., 278, 2011
|
|
3MVS
 
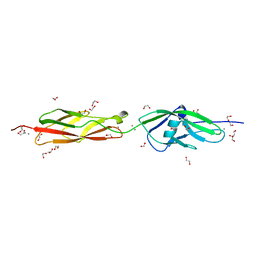 | | Structure of the N-terminus of Cadherin 23 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-23 | | Authors: | Clark, P, Joseph, J.S, Kolatkar, A.R. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1M5N
 
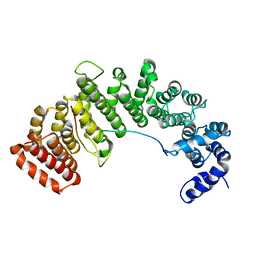 | | Crystal structure of HEAT repeats (1-11) of importin b bound to the non-classical NLS(67-94) of PTHrP | | Descriptor: | Importin beta-1 subunit, Parathyroid hormone-related protein | | Authors: | Cingolani, G, Bednenko, J, Gillespie, M.T, Gerace, L. | | Deposit date: | 2002-07-09 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis for the recognition
of a nonclassical nuclear localization
signal by importin beta
Mol.Cell, 10, 2002
|
|
3MWR
 
 | |
1M6D
 
 | | Crystal structure of human cathepsin F | | Descriptor: | 4-MORPHOLIN-4-YL-PIPERIDINE-1-CARBOXYLIC ACID [1-(3-BENZENESULFONYL-1-PROPYL-ALLYLCARBAMOYL)-2-PHENYLETHYL]-AMIDE, Cathepsin F | | Authors: | Somoza, J.R, Palmer, J.T, Ho, J.D. | | Deposit date: | 2002-07-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators
J.Mol.Biol., 322, 2002
|
|
3N3T
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
6SPO
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
3EKE
 
 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
4NRD
 
 | |
3EC7
 
 | | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2
To be Published
|
|
4NCY
 
 | | In situ trypsin crystallized on a MiTeGen micromesh with imidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6SR2
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 37 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
