2J9F
 
 | | Human branched-chain alpha-ketoacid dehydrogenase-decarboxylase E1b | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Jun, L, Machius, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2006-11-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Two Active Sites in Human Branched-Chain Alpha- Keto Acid Dehydrogenase Operate Independently without an Obligatory Alternating-Site Mechanism.
J.Biol.Chem., 282, 2007
|
|
5NYH
 
 | | Crystal Structure of Hsp90-alpha N-Domain in complex with Indazole derivative | | Descriptor: | Heat shock protein HSP 90-alpha, ~{N}-methyl-~{N}-(4-morpholin-4-ylphenyl)-6-oxidanyl-3-pyrrolidin-1-ylcarbonyl-2~{H}-indazole-5-carboxamide | | Authors: | Amaral, M. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5OCI
 
 | | Human Heat Shock Protein 90 bound to 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid methyl-(4-morpholin-4-yl-phenyl)-amide | | Descriptor: | 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid methyl-(4-morpholin-4-yl-phenyl)-amide, Heat shock protein HSP 90-alpha | | Authors: | Schuetz, D.A, Richter, L, Amaral, M, Grandits, M, Musil, D, Graedler, U, Frech, M, Ecker, G.F. | | Deposit date: | 2017-07-03 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5BUI
 
 | | ERK2 complexed with 2-pyridiyl tetrahydroazaindazole | | Descriptor: | 3-(4-fluorophenyl)-5-(pyridin-2-yl)-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase 1, NICKEL (II) ION | | Authors: | Bellamacina, C.R, Shu, W, Bussiere, D.E, Bagdanoff, J.T. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Ligand efficient tetrahydro-pyrazolopyridines as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2BF1
 
 | | Structure of an unliganded and fully-glycosylated SIV gp120 envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, B, Vogan, E.M, Gong, H, Skehel, J.J, Wiley, D.C, Harrison, S.C. | | Deposit date: | 2004-12-02 | | Release date: | 2005-02-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of an Unliganded Simian Immunodeficiency Virus Gp120 Core
Nature, 433, 2005
|
|
7CRH
 
 | | Cryo-EM structure of SKF83959 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1S)-6-chloranyl-3-methyl-1-(3-methylphenyl)-1,2,4,5-tetrahydro-3-benzazepine-7,8-diol, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yan, W, Shao, Z.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKW
 
 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
3W0D
 
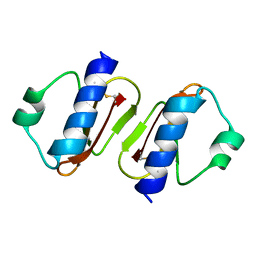 | | Structure of elastase inhibitor AFUEI (cyrstal form I) | | Descriptor: | Elastase inhibitor AFUEI | | Authors: | Imada, K, Sakuma, M, Okumura, Y, Ogawa, K, Nikai, T, Homma, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-15 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structure Analysis and Characterization of AFUEI, an Elastase Inhibitor from Aspergillus fumigatus
J.Biol.Chem., 288, 2013
|
|
3W0E
 
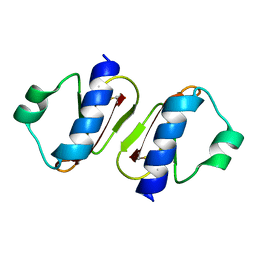 | | Structure of elastase inhibitor AFUEI (crystal form II) | | Descriptor: | Elastase inhibitor AFUEI | | Authors: | Imada, K, Sakuma, M, Okumura, Y, Ogawa, K, Nikai, T, Homma, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure Analysis and Characterization of AFUEI, an Elastase Inhibitor from Aspergillus fumigatus
J.Biol.Chem., 288, 2013
|
|
4IS8
 
 | |
2LK6
 
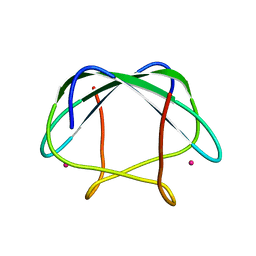 | | NMR determination of the global structure of the Cd-113 derivative of desulforedoxin | | Descriptor: | CADMIUM ION, Desulforedoxin | | Authors: | Goodfellow, B.J, Rusnak, F, Moura, I, Domke, T, Moura, J.J.G. | | Deposit date: | 2011-10-07 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the global structure of the 113Cd derivative of desulforedoxin: investigation of the hydrogen bonding pattern at the metal center.
Protein Sci., 7, 1998
|
|
4A4J
 
 | | Crosstalk between Cu(I) and Zn(II) homeostasis | | Descriptor: | COPPER-TRANSPORTING ATPASE PACS, ZINC ION | | Authors: | Badarau, A, Basle, A, Firbank, S.J, Denninson, C. | | Deposit date: | 2011-10-14 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigating the Role of Zinc and Copper Binding Motifs of Trafficking Sites in the Cyanobacterium Synechocystis Pcc 6803.
Biochemistry, 52, 2013
|
|
5HZX
 
 | | Crystal structure of zebrafish MTH1 in complex with TH588 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Gustafsson, R, Brautigam, L, Pudelko, L, Jemth, A.-S, Gad, H, Karsten, S, Carreras-Puigvert, J, Homan, E, Berndt, C, Berglund, U.W, Helleday, T, Stenmark, P. | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hypoxic Signaling and the Cellular Redox Tumor Environment Determine Sensitivity to MTH1 Inhibition.
Cancer Res., 76, 2016
|
|
1C48
 
 | | MUTATED SHIGA-LIKE TOXIN B SUBUNIT (G62T) | | Descriptor: | PROTEIN (SHIGA-LIKE TOXIN I B SUBUNIT) | | Authors: | Ling, H, Bast, D, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-08-11 | | Release date: | 2000-08-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the Primary Receptor Binding Site of Shiga-Like Toxin B Subunits: Structures of Mutated Shiga-Like Toxin I B-Pentamer with and without Bound Carbohydrate
To be Published
|
|
1C4Q
 
 | |
7N63
 
 | | X-ray structure of HCAN_0200, an aminotransferase from Helicobacter canadensis in complex with its external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Griffiths, W.A, Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7B
 
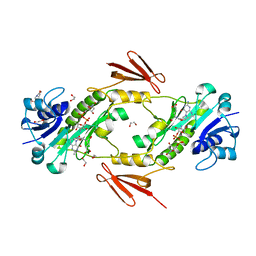 | | crystal structure of the N-formyltrasferase HCAN_0200 from Helicobacter canadensis on complex with folinic acid and dTDP-3-aminofucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
3QG7
 
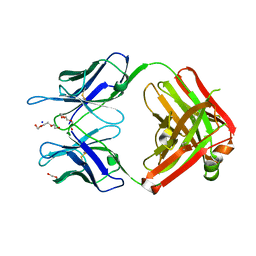 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Antibody Heavy Chain, AP4-24H11 Antibody Light Chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Kirchdoerfer, R.K, Kaufmann, G.F, Janda, J.D, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
2IYK
 
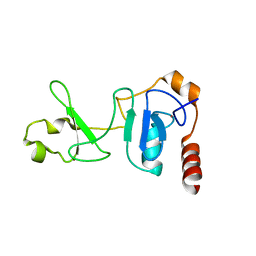 | | Crystal structure of the UPF2-interacting domain of nonsense mediated mRNA decay factor UPF1 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, ZINC ION | | Authors: | Kadlec, J, Guilligay, D, Ravelli, R.B, Cusack, S. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Upf2-Interacting Domain of Nonsense-Mediated Mrna Decay Factor Upf1.
RNA, 12, 2006
|
|
4A48
 
 | | Crosstalk between Cu(I) and Zn(II) homeostasis | | Descriptor: | MAGNESIUM ION, PROBABLE COPPER-TRANSPORTING ATPASE PACS, ZINC ION | | Authors: | Badarau, A, Basle, A, Firbank, S.J, Denninson, C. | | Deposit date: | 2011-10-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Investigating the Role of Zinc and Copper Binding Motifs of Trafficking Sites in the Cyanobacterium Synechocystis Pcc 6803.
Biochemistry, 52, 2013
|
|
3UFL
 
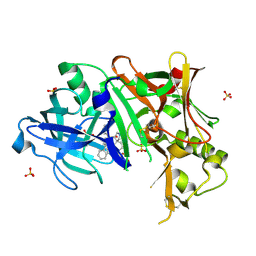 | | Discovery of Pyrrolidine-based b-Secretase Inhibitors: Lead Advancement through Conformational Design for Maintenance of Ligand Binding Efficiency | | Descriptor: | (1R,4'S)-3,4-dihydro-2H-spiro[naphthalene-1,3'-pyrrolidin]-4'-yl[(2S,4R)-2,4-diphenylpiperidin-1-yl]methanone, Beta-secretase 1, GLYCEROL, ... | | Authors: | Allison, T, Munshi, S, Soisson, S.M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of pyrrolidine-based beta-secretase inhibitors: Lead advancement through conformational design for maintenance of ligand binding efficiency.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1RAE
 
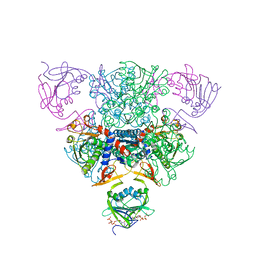 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
