1KWR
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH INHIBITOR 0134-36 | | Descriptor: | 1-METHYL-3-OXO-1,3-DIHYDRO-BENZO[C]ISOTHIAZOLE-5-SULFONIC ACID AMIDE, Carbonic anhydrase II, ZINC ION | | Authors: | Grueneberg, S, Stubbs, M.T. | | Deposit date: | 2002-01-30 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Successful virtual screening for novel inhibitors of human carbonic anhydrase: strategy and experimental confirmation.
J.Med.Chem., 45, 2002
|
|
4Z84
 
 | | PKAB3 in complex with pyrrolidine inhibitor 34a | | Descriptor: | 7-[(3S,4R)-4-(3-chlorophenyl)carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, METHANOL, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Lund, B.A, Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of PKA and a PKB Mimic.
Chemistry, 22, 2016
|
|
4ZBO
 
 | | Streptomyces bingchenggensis acetoacetate decarboxylase in non-covalent complex with potassium formate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acetoacetate decarboxylase, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sbi00515, a Protein of Unknown Function from Streptomyces bingchenggensis, Highlights the Functional Versatility of the Acetoacetate Decarboxylase Scaffold.
Biochemistry, 54, 2015
|
|
1K2N
 
 | |
3MR1
 
 | |
4ZBT
 
 | | Streptomyces bingchenggensis aldolase-dehydratase in Schiff base complex with pyruvate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acetoacetate decarboxylase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sbi00515, a Protein of Unknown Function from Streptomyces bingchenggensis, Highlights the Functional Versatility of the Acetoacetate Decarboxylase Scaffold.
Biochemistry, 54, 2015
|
|
4ZGM
 
 | | Crystal structure of Semaglutide peptide backbone in complex with the GLP-1 receptor extracellular domain | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, Glucagon-like peptide 1 receptor, Semaglutide peptide backbone; 8Aib,34R-GLP-1(7-37)-OH | | Authors: | Reedtz-Runge, S. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Once-Weekly Glucagon-Like Peptide-1 (GLP-1) Analogue Semaglutide.
J.Med.Chem., 58, 2015
|
|
1QYV
 
 | |
4Z83
 
 | | PKAB3 in complex with pyrrolidine inhibitor 47a | | Descriptor: | 7-{(3S,4R)-4-[(5-bromothiophen-2-yl)carbonyl]pyrrolidin-3-yl}quinazolin-4(3H)-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Lund, B.A, Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of PKA and a PKB Mimic.
Chemistry, 22, 2016
|
|
1TEC
 
 | | CRYSTALLOGRAPHIC REFINEMENT BY INCORPORATION OF MOLECULAR DYNAMICS. THE THERMOSTABLE SERINE PROTEASE THERMITASE COMPLEXED WITH EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, SODIUM ION, ... | | Authors: | Gros, P, Dijkstra, B.W, Hol, W.G.J. | | Deposit date: | 1989-05-24 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic refinement by incorporation of molecular dynamics: thermostable serine protease thermitase complexed with eglin c.
Acta Crystallogr.,Sect.B, 45, 1989
|
|
3MX6
 
 | |
4A04
 
 | | Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome | | Descriptor: | DNA, T-BOX TRANSCRIPTION FACTOR TBX1 | | Authors: | El Omari, K, De Mesmaeker, J, Karia, D, Ginn, H, Bhattacharya, S, Mancini, E.J. | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the DNA-Bound T-Box Domain of Human Tbx1, a Transcription Factor Associated with the Digeorge Syndrome
Proteins, 80, 2012
|
|
4ZZX
 
 | | Structure of PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
3C2Y
 
 | | tRNA-Guanine Transglycosylase (TGT) in complex with 6-Amino-2-methyl-1,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-methyl-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2008-01-26 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
1WPD
 
 | | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins | | Descriptor: | Potassium channel toxin alpha-KTx 6.2,Potassium channel toxin alpha-KTx 6.3 | | Authors: | Regaya, I, Beeton, C, Ferrat, G, Andreotti, N, Chandy, G.K, Darbon, H, De Waard, M, Sabatier, J.M. | | Deposit date: | 2004-09-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins
J.Biol.Chem., 279, 2004
|
|
1WMM
 
 | |
3CM2
 
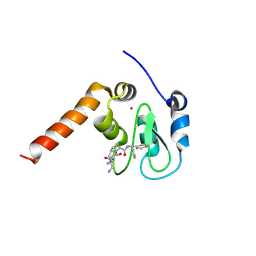 | | Crystal Structure of XIAP BIR3 domain in complex with a Smac-mimetic compound, Smac010 | | Descriptor: | (3S,6S,7R,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-7-(aminomethyl)-N-(diphenylmethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Cossu, F, Mastrangelo, E, Milani, M. | | Deposit date: | 2008-03-20 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane smac mimetics. Structure, activity, and recognition principles.
J.Mol.Biol., 384, 2008
|
|
3CM7
 
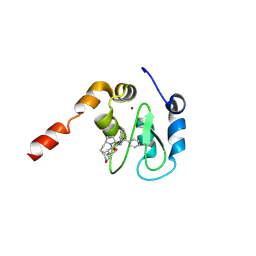 | | Crystal Structure of XIAP-BIR3 domain in complex with Smac-mimetic compuond, Smac005 | | Descriptor: | (3S,6S,7S,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-N-(diphenylmethyl)-7-(hydroxymethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Mastrangelo, E, Cossu, F, Milani, M. | | Deposit date: | 2008-03-21 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane smac mimetics. Structure, activity, and recognition principles.
J.Mol.Biol., 384, 2008
|
|
3CWB
 
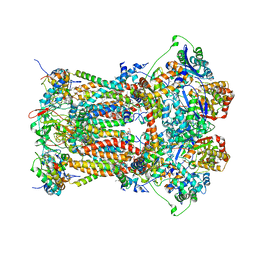 | | Chicken Cytochrome BC1 Complex inhibited by an iodinated analogue of the polyketide Crocacin-D | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L, Cromartie, T, Viner, R, Crowley, P.J, Berry, E.A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D.
Bioorg.Med.Chem., 16, 2008
|
|
2BO9
 
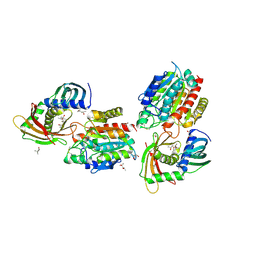 | | Human carboxypeptidase A4 in complex with human latexin. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONE, ... | | Authors: | Pallares, I, Bonet, R, Garcia-Castellanos, R, Ventura, S, Aviles, F.X, Vendrell, J, Gomis-Rueth, F.X. | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Human Carboxypeptidase A4 with its Endogenous Protein Inhibitor, Latexin.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7AWR
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tegafur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, TEGAFUR | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AX6
 
 | | Structure of SARS-CoV-2 Main Protease bound to Glutathione isopropyl ester | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AY7
 
 | | Structure of SARS-CoV-2 Main Protease bound to Isofloxythepin | | Descriptor: | 3C-like proteinase, 9-fluoranyl-3-propan-2-yl-5,6-dihydrobenzo[b][1]benzothiepine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AZU
 
 | |
7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
