6WPJ
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and d4T | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA Primer 21-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Molecules, 25, 2020
|
|
6X4D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | Descriptor: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6WPH
 
 | |
6X4B
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-5-fluoro-8-methyl-2-naphthonitrile (JLJ655), a Non-nucleoside Inhibitor | | Descriptor: | 7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-5-fluoro-8-methylnaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6WB0
 
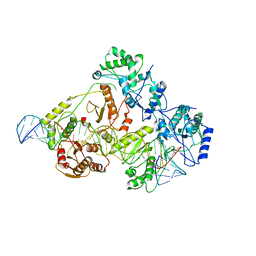 | | +3 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WB2
 
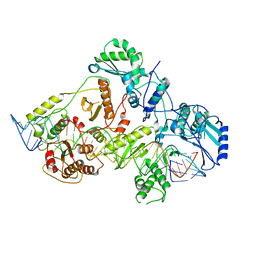 | | +3 extended HIV-1 reverse transcriptase initiation complex core (displaced state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
7SEP
 
 | |
7SLS
 
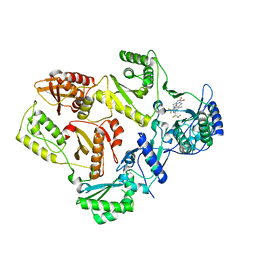 | | HIV Reverse Transcriptase with compound Pyr02 | | Descriptor: | 5-(difluoromethyl)-3-{[1-{[(3S)-5-fluoro-2-methyl-6-oxo-3,6-dihydropyridin-3-yl]methyl}-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl]oxy}-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
7SLR
 
 | | HIV Reverse Transcriptase with compound Pyr01 | | Descriptor: | 5-(difluoromethyl)-3-({1-[(5-fluoro-2-oxo-1,2-dihydropyridin-3-yl)methyl]-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl}oxy)-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
7SJX
 
 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
7TAZ
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with VM-1500A, a non-nucleoside RT inhibitor | | Descriptor: | 2-[4-bromo-3-(3-chloro-5-cyanophenoxy)-2-fluorophenyl]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase p51, Reverse transcriptase/ribonuclease H | | Authors: | Snyder, A.A, Risener, C.J, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2021-12-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with VM-1500A, a non-nucleoside RT inhibitor
To Be Published
|
|
7U5Z
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with JLJ353 | | Descriptor: | 2-chloro-4-({5-[(2,6-difluorophenyl)methyl]-1,3-oxazol-2-yl}amino)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological testing of biphenylmethyloxazole inhibitors targeting HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
7UY5
 
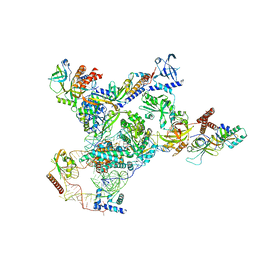 | | Tetrahymena telomerase with CST | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Song, H, Chan, H, Wang, Y, Liu, B, Susac, L, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase.
Nature, 608, 2022
|
|
7UY6
 
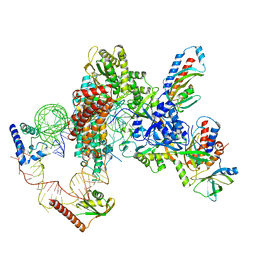 | | Tetrahymena telomerase at 2.9 Angstrom resolution | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Song, H, Chan, H, Wang, Y, Liu, B, Susac, L, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase.
Nature, 608, 2022
|
|
7SR6
 
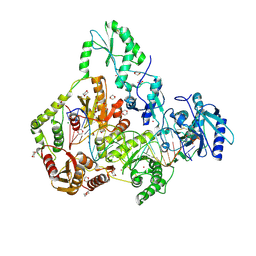 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNZ
 
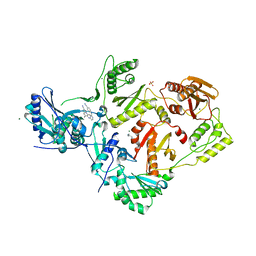 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((2-((4-cyanophenyl)amino)pyrimidin-4-yl)amino)-5,7-dimethylindolizine-2-carbonitrile (JLJ604) | | Descriptor: | (4R)-6-{[2-(4-cyanoanilino)pyrimidin-4-yl]amino}-5,7-dimethylindolizine-2-carbonitrile, 1,4-DIAMINOBUTANE, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO2
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO6
 
 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO1
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.727 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SNP
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-(2-morpholinoethoxy)phenoxy)phenyl)acrylonitrile (JLJ530) | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-[2-(morpholin-4-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO4
 
 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase K103N/Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.767 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
1IKX
 
 | |
1IKV
 
 | | K103N Mutant HIV-1 Reverse Transcriptase in Complex with Efivarenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1J5O
 
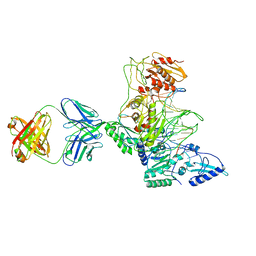 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
