6IO0
 
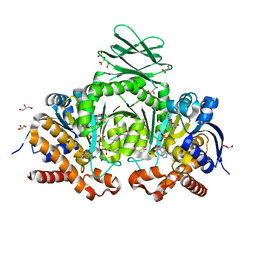 | | Human IDH1 R132C mutant complexed with compound A. | | Descriptor: | (2E)-3-{3-[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazole-4-carbonyl]-1-methyl-1H-indol-7-yl}prop-2-enoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Suzuki, M, Baba, D, Hanzawa, H. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Potent Blood-Brain Barrier-Permeable Mutant IDH1 Inhibitor Suppresses the Growth of Glioblastoma with IDH1 Mutation in a Patient-Derived Orthotopic Xenograft Model.
Mol.Cancer Ther., 19, 2020
|
|
5TWP
 
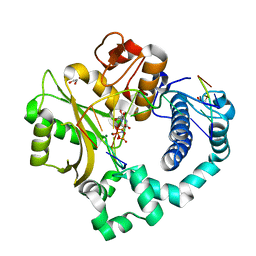 | | Pre-catalytic ternary complex of human Polymerase Mu with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
3HY2
 
 | | Crystal Structure of Sulfiredoxin in Complex with Peroxiredoxin I and ATP:Mg2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxiredoxin-1, ... | | Authors: | Jonsson, T.J, Johnson, L.C, Lowther, W.T. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Engineering of the Quaternary Sulfiredoxin-Peroxiredoxin Enzyme-Substrate Complex Reveals the Molecular Basis for Cysteine Sulfinic Acid Phosphorylation
J.Biol.Chem., 284, 2009
|
|
7LT0
 
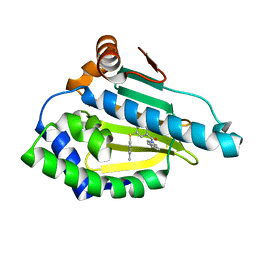 | | Hsp90a N-terminal inhibitor | | Descriptor: | (1,3-dihydro-2H-isoindol-2-yl){3-[(3,4-dimethylphenyl)sulfanyl]-4-hydroxyphenyl}methanone, Heat shock protein HSP 90-alpha | | Authors: | Balch, M, Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6MI6
 
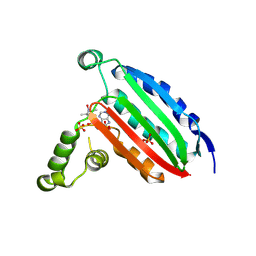 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH AN ADP ANALOG | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(1-hydroxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl]disulfanyl}phosphoryl]oxy}phosphoryl]adenosine, ADENOSINE-5'-DIPHOSPHATE, Chemotaxis protein CheA, ... | | Authors: | Crane, B.R, Muok, A.R, Chua, T.K, Le, H. | | Deposit date: | 2018-09-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Nucleotide Spin Labeling for ESR Spectroscopy of ATP-Binding Proteins.
Appl.Magn.Reson., 49, 2018
|
|
5VWC
 
 | | Crystal structure of human Scribble PDZ1 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein scribble homolog | | Authors: | Lim, K.Y.B, Kvansakul, M. | | Deposit date: | 2017-05-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor beta-PIX.
J. Biol. Chem., 292, 2017
|
|
5AC7
 
 | | S. enterica HisA with mutations D7N, D10G, dup13-15 | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SODIUM ION, ... | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5AHF
 
 | | Crystal structure of Salmonella enterica HisA D7N with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
6CCP
 
 | |
6CCV
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Rifampicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
5XJ6
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the glycerol 3-phosphate form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5B2Y
 
 | | Crystal Structure of P450BM3 with N-perfluorodecanoyl-L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,7,7,8,8,9,9,10,10,10-nonadecakis(fluoranyl)decanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
6APV
 
 | |
6G6Y
 
 | | Eg5-inhibitor complex | | Descriptor: | (~{N}~{Z})-~{N}-[(5~{S})-4-ethanoyl-5-methyl-5-phenyl-1,3,4-thiadiazolidin-2-ylidene]ethanamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F, Tham, C.L. | | Deposit date: | 2018-04-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Eg5 - K858 complex and implications for structure-based design of thiadiazole-containing inhibitors.
Eur J Med Chem, 156, 2018
|
|
3HHS
 
 | | Crystal Structure of Manduca sexta prophenoloxidase | | Descriptor: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | Authors: | Li, Y, Wang, Y, Jiang, H, Deng, J. | | Deposit date: | 2009-05-17 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5GWJ
 
 | | Structure of a Human topoisomerase IIbeta fragment in complex with DNA and E7873S | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Wang, Y.R, Chen, S.F, Wu, C.C, Chan, N.L. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry
Nucleic Acids Res., 45, 2017
|
|
6FNF
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with NVP-BHG712 | | Descriptor: | 4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
5B2U
 
 | | Crystal Structure of P450BM3 with N-perfluorohexanoyl -L-tryptophan | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[2,2,3,3,4,4,5,5,6,6,6-undecakis(fluoranyl)hexanoylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Cong, Z, Shoji, O, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of P450BM3 with decoy molecules
to be published
|
|
6LRM
 
 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
5ICC
 
 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, POTASSIUM ION, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
5TUS
 
 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U64
 
 | | Camel nanobody VHH-28 | | Descriptor: | VHH-28 | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers.
Sci Rep, 7, 2017
|
|
5Z3K
 
 | |
6MS1
 
 | | Crystal structure of the human Scribble PDZ1 domain bound to the PDZ-binding motif of APC | | Descriptor: | 1,2-ETHANEDIOL, APC C-terminus peptide, Protein scribble homolog | | Authors: | How, J.Y, Kvansakul, M, Caria, S, Humbert, P.O. | | Deposit date: | 2018-10-16 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the human Scribble PDZ1 domain bound to the PDZ-binding motif of APC.
FEBS Lett., 593, 2019
|
|
