7SMD
 
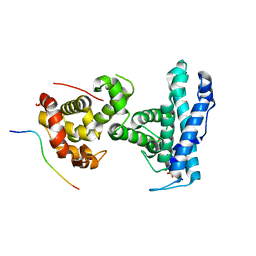 | | p107 pocket domain complexed with EID1 peptide | | Descriptor: | EP300-interacting inhibitor of differentiation 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7RUP
 
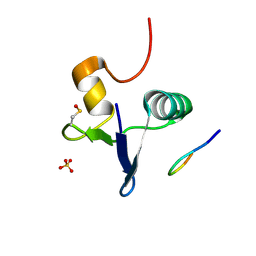 | | Structure of the human GIGYF2-TNRC6A complex | | Descriptor: | GRB10-interacting GYF protein 2, SULFATE ION, Trinucleotide repeat-containing gene 6A protein | | Authors: | Sobti, M, Mead, B.J, Igreja, C, Stewart, A.G, Christie, M. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Molecular basis for GIGYF-TNRC6 complex assembly.
Rna, 29, 2023
|
|
7TDR
 
 | |
7TDS
 
 | |
4XW2
 
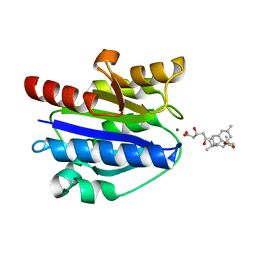 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | Descriptor: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | Authors: | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2015-01-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
4NXQ
 
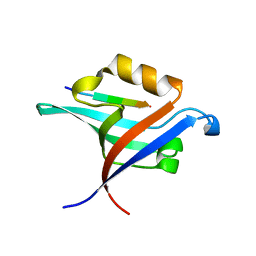 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Caspr4 Peptide | | Descriptor: | Contactin-associated protein-like 4 peptide, T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
7Z1K
 
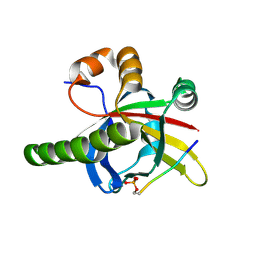 | | Crystal structure of the SPOC domain of human SHARP (SPEN) in complex with RNA polymerase II CTD heptapeptide phosphorylated on Ser5 | | Descriptor: | Msx2-interacting protein, SER-TYR-SER-PRO-THR-SEP | | Authors: | Appel, L, Grishkovskaya, I, Slade, D, Djinovic-Carugo, K. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators.
Nat Commun, 14, 2023
|
|
7YNK
 
 | |
7YNJ
 
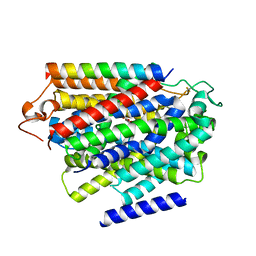 | |
4ZEQ
 
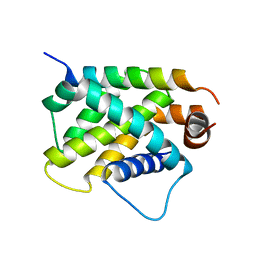 | | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide, Northeast Structural Genomics Consortium Target HX9247 | | Descriptor: | BH3-interacting domain death agonist, Bcl-2-related protein A1 | | Authors: | Guan, R, Xiao, R, Mao, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of human BFL-1 in complex with tBid BH3 peptide
To Be Published
|
|
8GLC
 
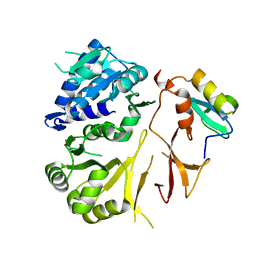 | |
6DCD
 
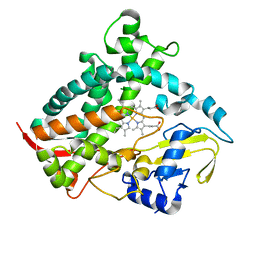 | |
4H8S
 
 | | Crystal structure of human APPL2BARPH domain | | Descriptor: | DCC-interacting protein 13-beta | | Authors: | Martin, J.L, King, G.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane Curvature Protein Exhibits Interdomain Flexibility and Binds a Small GTPase.
J.Biol.Chem., 287, 2012
|
|
6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
6N6R
 
 | |
1FEX
 
 | |
7KSB
 
 | | Crystal structure on Act c 10.0101 | | Descriptor: | Non-specific lipid-transfer protein 1, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Giangrieco, I, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
8XHO
 
 | |
7LY5
 
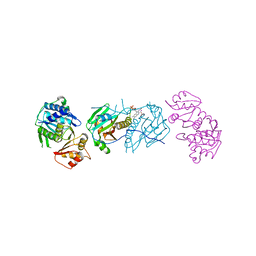 | | Proteolyzed crystal structure of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | BmdB, Bacillamide NRPS, BmdC, ... | | Authors: | Fortinez, C.M, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
7LY4
 
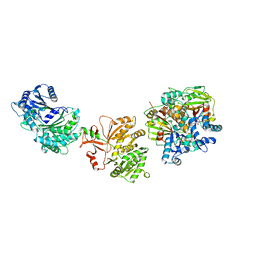 | | Cryo-EM structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase, BmdC | | Descriptor: | BmdB, bacillamide NRPS, BmdC, ... | | Authors: | Sharon, I, Fortinez, C.M, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
7LY7
 
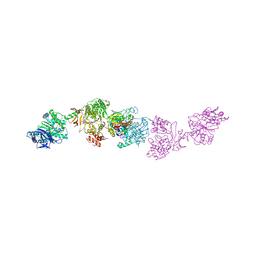 | | Crystal structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | 5'-{[(2R,3S)-3-amino-2-({2-[(N-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-4-sulfanylbutane-1-sulfonyl]amino}-5'-deoxyadenosine, BmdB, Bacillamide NRPS, ... | | Authors: | Fortinez, C.M, Sharon, I, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4BP9
 
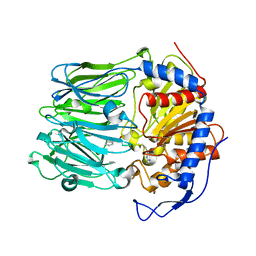 | | Oligopeptidase B from Trypanosoma brucei with covalently bound antipain - closed form | | Descriptor: | ANTIPAIN, OLIGOPEPTIDASSE B | | Authors: | Canning, P, Rea, D, Morty, R, Fulop, V. | | Deposit date: | 2013-05-23 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of Trypanosoma Brucei Oligopeptidase B Broaden the Paradigm of Catalytic Regulation in Prolyl Oligopeptidase Family Enzymes.
Plos One, 8, 2013
|
|
4X9U
 
 | | Crystal structure of the kiwifruit allergen Act d 5 | | Descriptor: | Kiwellin | | Authors: | Offermann, L.R, Perdue, M.L, Giangrieco, I, Tamburrini, M, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elusive Structural, Functional, and Immunological Features of Act d 5, the Green Kiwifruit Kiwellin.
J.Agric.Food Chem., 63, 2015
|
|
3JC2
 
 | |
