6P0D
 
 | | Human DNA Ligase 1 (E346A/E592A) Bound to an Adenylated, hydroxyl terminated DNA nick | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6P0C
 
 | | Human DNA Ligase 1 Bound to an Adenylated, hydroxyl terminated DNA nick in EDTA | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Riccio, A.A, Williams, R.S. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6P09
 
 | | Human DNA Ligase 1 Bound to an Adenylated, dideoxy Terminated DNA nick with 200 mM Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6F5N
 
 | | Nickel-Bound Crystal Structure of a GB1 Variant | | Descriptor: | NICKEL (II) ION, Nickel-Binding Protein | | Authors: | Rothlisberger, U, Bozkurt, E, Hovius, R, Perez, M.A.S, Browning, N.J. | | Deposit date: | 2017-12-01 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nickel-Mediated Self-Assembly of an Ultrastable GB1 Variant
To Be Published
|
|
6P0E
 
 | | Human DNA Ligase 1 (E346A,E592A) bound to adenylated DNA containing an 8-oxo guanine:adenine base-pair | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6FPJ
 
 | | Structure of the AMPAR GluA3 N-terminal domain bound to phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
6FOG
 
 | |
6FOH
 
 | |
6G1L
 
 | | MITF/CLEARbox structure | | Descriptor: | CLEAR-box, Microphthalmia-associated transcription factor | | Authors: | Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-03-21 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MITF has a central role in regulating starvation-induced autophagy in melanoma.
Sci Rep, 9, 2019
|
|
6FYM
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK1 | | Descriptor: | 7,8-dimethyl-2-(pyrimidin-2-ylsulfanylmethyl)-3~{H}-quinazolin-4-one, NITRATE ION, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | Deposit date: | 2018-03-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
4ABK
 
 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 IN COMPLEX WITH ADENOSINE- 5-DIPHOSPHORIBOSE | | Descriptor: | POLY [ADP-RIBOSE] POLYMERASE 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
7K4D
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 3OG | | Descriptor: | 5-[(4-{trans-4-hydroxy-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4B
 
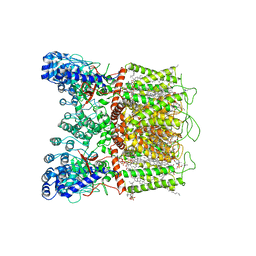 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor cis-22a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[cis-4-(3-methylphenyl)cyclohexyl]-4-(pyridin-3-yl)piperazine, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
6Q1V
 
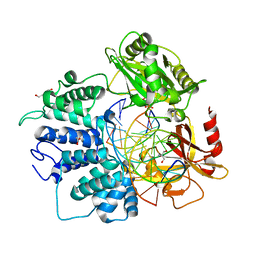 | | Human DNA Ligase 1 (E592R) Bound to an Adenylated, hydroxyl terminated DNA nick | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-08-06 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6Q8P
 
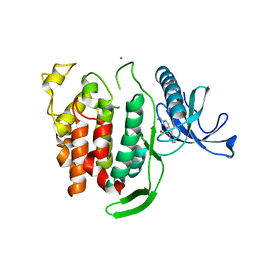 | | Structure of CLK1 with bound N-methyl-10-nitropyrido[3,4-g]quinazolin-2-amine | | Descriptor: | Dual specificity protein kinase CLK1, POTASSIUM ION, ~{N}-methyl-10-nitro-pyrido[3,4-g]quinazolin-2-amine | | Authors: | Joerger, A.C, Chatterjee, D, Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Josselin, B, Baratte, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
7K4E
 
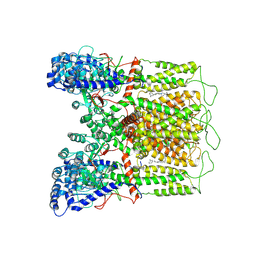 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 30 | | Descriptor: | 5-({4-[(1R,4S)-3'-methyl[1,2,3,4-tetrahydro[1,1'-biphenyl]]-4-yl]piperazin-1-yl}methyl)pyridin-2(1H)-one, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
3PDH
 
 | |
4ABL
 
 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 | | Descriptor: | BROMIDE ION, POLY [ADP-RIBOSE] POLYMERASE 14 | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
4AE5
 
 | |
4AM6
 
 | | C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, SULFATE ION | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AM7
 
 | | ADP-BOUND C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZUG
 
 | | E268D mutant of FAD synthetase from Corynebacterium ammoniagenes | | Descriptor: | RIBOFLAVIN BIOSYNTHESIS PROTEIN RIBF, SULFATE ION | | Authors: | Herguedas, B, Martinez-Julvez, M, Serrano, A, Medina, M. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Key Residues at the Riboflavin Kinase Catalytic Site of the Bifunctional Riboflavin Kinase/Fmn Adenylyltransferase from Corynebacterium Ammoniagenes.
Cell Biochem.Biophys., 65, 2013
|
|
6QU7
 
 | | Crystal structure of human DHODH in complex with BAY 2402234 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Friberg, A, Gradl, S. | | Deposit date: | 2019-02-26 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies.
Leukemia, 33, 2019
|
|
7K4C
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromopyridin-3-yl)-4-[cis-4-(3-methylphenyl)cyclohexyl]piperazine, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
6FZM
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK6 | | Descriptor: | 4-[(8-methyl-4-oxidanylidene-7-prop-1-ynyl-3~{H}-quinazolin-2-yl)methylsulfanyl]benzoic acid, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
