8UFU
 
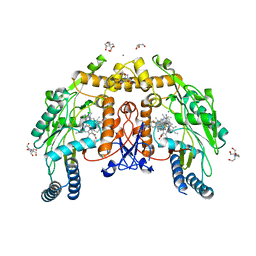 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(9-amino-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)-4-methylquinolin-2-amine | | Descriptor: | (7M)-7-[(9S)-9-amino-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl]-4-methylquinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
4N75
 
 | |
8U8K
 
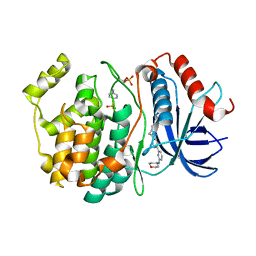 | |
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
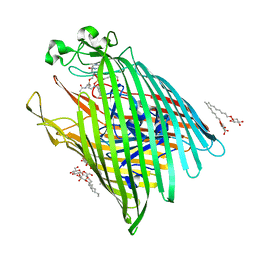
6E4V
 
 | |
8UPP
 
 | | Campylobacter jejuni ketol-acid reductoisomerase in complex with NADPH and Hoe704 | | Descriptor: | (2R)-(dimethylphosphoryl)(hydroxy)acetic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Lin, X, Lv, Y, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2023-10-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mapping of the Reaction Trajectory catalyzed by Class I Ketol-Acid Reductoisomerase
Acs Catalysis, 2024
|
|
8UD5
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
8G1Z
 
 | |
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
8G20
 
 | |
4MRH
 
 | |
8UFP
 
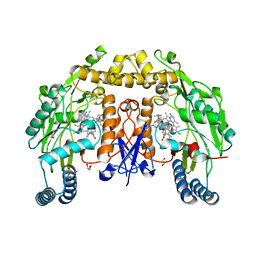 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-methyl-7-(4-methyl-2,3,4,5-tetrahydrobenzo[f][1,4]oxazepin-7-yl)quinolin-2-amine dihydrochloride | | Descriptor: | (7M)-4-methyl-7-(4-methyl-2,3,4,5-tetrahydro-1,4-benzoxazepin-7-yl)quinolin-2-amine, GLYCEROL, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
4MU1
 
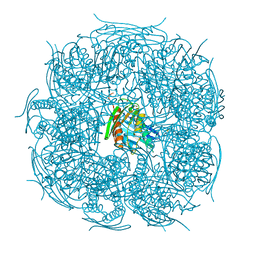 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+, imidazole, and sulfate at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
6E88
 
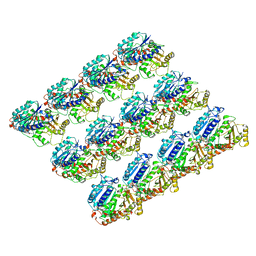 | | Cryo-EM structure of C. elegans GDP-microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha-2 chain, ... | | Authors: | Chaaban, S, Jariwala, S, Chieh-Ting, H, Redemann, S, Kollman, J, Muller-Reichert, T, Sept, D, Bui, K.H, Brouhard, G.J. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth.
Dev. Cell, 47, 2018
|
|
8UD4
 
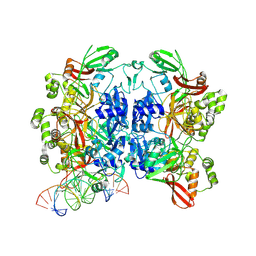 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
4MV8
 
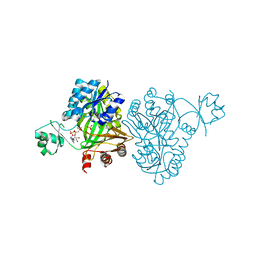 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with AMPPCP and Phosphate | | Descriptor: | Biotin carboxylase, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MUG
 
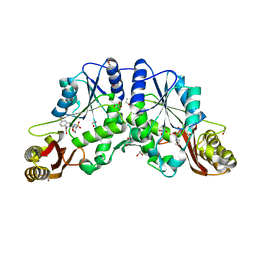 | |
4MVX
 
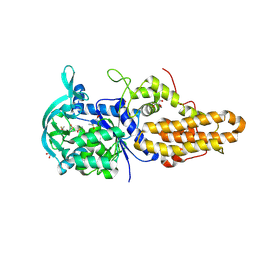 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-phenylurea (Chem 1356) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-phenylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MUX
 
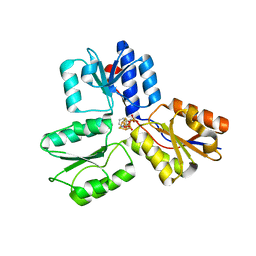 | | IspH in complex with pyridin-3-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, pyridin-3-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
4MWC
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[(2-methyl-1-benzothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1540) | | Descriptor: | 1-(3-{[(2-methyl-1-benzothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MWZ
 
 | |
6E23
 
 | |
8UD2
 
 | | SARS-CoV-2 Nsp15, apo-form | | Descriptor: | Non-structural protein 15 | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
6E22
 
 | |
4MYV
 
 | | Free HSV-2 gD structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
