6NBI
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
4UTM
 
 | | XenA - Reduced - Y183F variant in complex with 8-hydroxycoumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 8-HYDROXYCOUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UUM
 
 | |
4UX6
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4UTO
 
 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4UVB
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1S,2R) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,7aS)-1-amino-1,10,11-trimethyl-4,6-dioxo-3-phenyl-2,3,5,6,7,7a-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
3M3E
 
 | |
4UWD
 
 | |
4URS
 
 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
4USW
 
 | | Crystal structure of human soluble Adenylyl Cyclase with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2014-07-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of Human Soluble Adenylyl Cyclase and Crystal Structures of its Nucleotide Complexes -Implications for Cyclase Catalysis and Evolution.
FEBS J., 281, 2014
|
|
4UXN
 
 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
4V37
 
 | |
6NUC
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
4UTS
 
 | | Room temperature crystal structure of the fast switching M159T mutant of fluorescent protein Dronpa | | Descriptor: | FLUORESCENT PROTEIN DRONPA | | Authors: | Kaucikas, M, Fitzpatrick, A, Bryan, E, Struve, A, Henning, R, Kosheleva, I, Srajer, V, van Thor, J.J. | | Deposit date: | 2014-07-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Room Temperature Crystal Structure of the Fast Switching M159T Mutant of the Fluorescent Protein Dronpa.
Proteins, 83, 2015
|
|
4V0N
 
 | |
4V1M
 
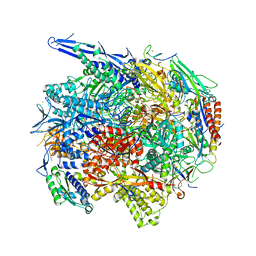 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4UYH
 
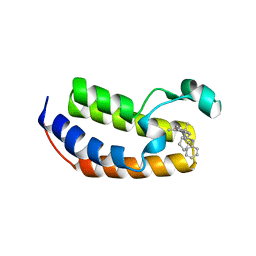 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1-((2R,4S)-2-methyl-4-(phenylamino)-6-(4-(piperidin-1-ylmethyl)phenyl)-3,4-dihydroquinolin-1(2H)-yl)ethanone | | Descriptor: | 1-[(2S,4R)-2-methyl-4-(phenylamino)-6-[4-(piperidin-1-ylmethyl)phenyl]-3,4-dihydroquinolin-1(2H)-yl]ethanone, BROMODOMAIN-CONTAINING PROTEIN 2, DIMETHYL SULFOXIDE, ... | | Authors: | Chung, C, Bamborough, P, Gosmini, R. | | Deposit date: | 2014-08-31 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Discovery of I-Bet726 (Gsk1324726A), a Potent Tetrahydroquinoline Apoa1 Up-Regulator and Selective Bet Bromodomain Inhibitor.
J.Med.Chem., 57, 2014
|
|
4UVC
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Phenyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-[5-[(1S)-1-azanyl-1,3-diphenyl-propyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-4aH-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UTK
 
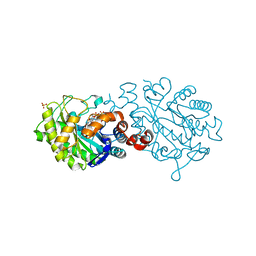 | | XenA - reduced - Y183F variant | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, SULFATE ION, XENOBIOTIC REDUCTASE | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UUN
 
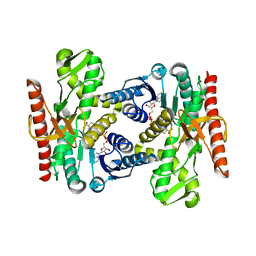 | |
6SGP
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a sulfamide inhibitor GluGlu | | Descriptor: | (2~{S})-2-[[(2~{S})-1,5-bis(oxidanyl)-1,5-bis(oxidanylidene)pentan-2-yl]sulfamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Shukla, S, Motlova, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
4WA6
 
 | |
6NBH
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
4URA
 
 | | Crystal structure of human JMJD2A in complex with compound 14a | | Descriptor: | 1,2-ETHANEDIOL, 2-(2H-1,2,3-triazol-4-yl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Krojer, T, England, K.S, Vollmar, M, Crawley, L, Williams, E, Riesebos, E, Szykowska, A, Burgess-Brown, N, Oppermann, U, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor.
Medchemcomm, 5, 2014
|
|
4UTL
 
 | | XenA - reduced - Y183F variant in complex with coumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, COUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.229 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
