8FOR
 
 | |
8TFX
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with 2',5'-ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-2'-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6F4P
 
 | | Human JMJD5 in complex with MN, NOG and RPS6 (129-144) (complex-1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 40S ribosomal protein S6, JmjC domain-containing protein 5, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6F6T
 
 | |
4LE0
 
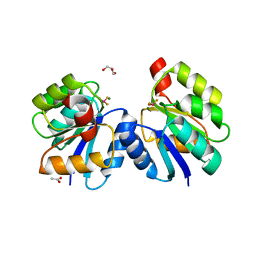 | | Crystal structure of the receiver domain of DesR in complex with beryllofluoride and magnesium | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
7OR4
 
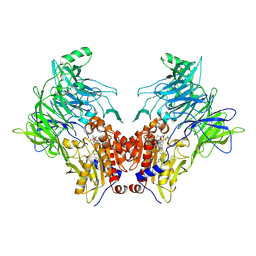 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, B142 | | Descriptor: | Dipeptidyl peptidase 8, methyl 3-[4-[(4-bromophenyl)methyl]piperazin-1-yl]carbonyl-5-[(2-ethyl-2-methanoyl-butanoyl)amino]benzoate, trimethylamine oxide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8TFY
 
 | |
7OLE
 
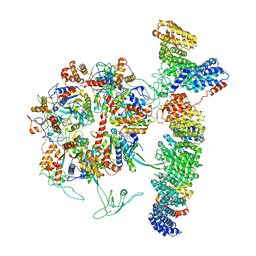 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
7OZ7
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, L84 | | Descriptor: | Dipeptidyl peptidase 8, trimethylamine oxide, ~{N}-[3-[[4-[(4-bromophenyl)methyl]piperazin-1-yl]methyl]phenyl]-2-ethyl-2-methanoyl-butanamide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6F6B
 
 | |
7OW0
 
 | |
7OXS
 
 | |
8FTA
 
 | |
6F9K
 
 | |
4LIW
 
 | |
6F8L
 
 | |
8FO9
 
 | |
7ORF
 
 | | Crystal structure of JNK3 in complex with FMU-001-367 (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 10, ... | | Authors: | Chaikuad, A, Koch, P, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8TRG
 
 | |
4LO0
 
 | | Apo HA17-HA33 | | Descriptor: | HA-17, HA-33 | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
8TNS
 
 | |
4LAP
 
 | |
7P18
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Sterolibacterium denitrificans in complex with 1,4-androstadiene-3,17-dione | | Descriptor: | 3-oxosteroid 1-dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wojcik, P, Mrugala, B, Kurpiewska, K, Szaleniec, M. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure, Mutagenesis, and QM:MM Modeling of 3-Ketosteroid Delta 1 -Dehydrogenase from Sterolibacterium denitrificans ─The Role of a New Putative Membrane-Associated Domain and Proton-Relay System in Catalysis.
Biochemistry, 62, 2023
|
|
4LNF
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-Q | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
7P7X
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (holo form). | | Descriptor: | ACETATE ION, Aminotransferase class IV, PHOSPHATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis : Biochemical and Structural Insights into the New Enzyme.
Molecules, 26, 2021
|
|
