6CMM
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine inhibitor
To Be Published
|
|
2GPQ
 
 | |
6D3A
 
 | | Structure of human ARH3 D314E bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.60001016 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YOR
 
 | |
6CSW
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor | | Descriptor: | (7R)-5-butyl-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7,8-dimethyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Chiodi, C.G, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-methyl-N-propyl-dihydropteridine inhibitor
To Be Published
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2F4U
 
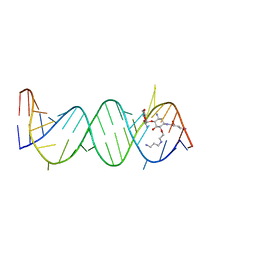 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-((1R,2S,3R,4R,5S)-5-AMINO-4-[(2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-{2-[(3-AMINOPROPYL)AMINO]ETHOXY}-3-HYDROXYCYCLOHEXYL)-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
4YOU
 
 | |
6EM1
 
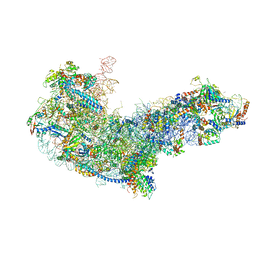 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
2LRQ
 
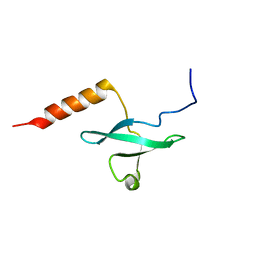 | | Chemical Shift Assignment and Solution Structure of Fr822A from Drosophila melanogaster. Northeast Structural Genomics Consortium Target Fr822A | | Descriptor: | NuA4 complex subunit EAF3 homolog | | Authors: | Lee, H, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2012-04-11 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Fr822A from Drosophila melanogaster.
To be Published
|
|
1HNX
 
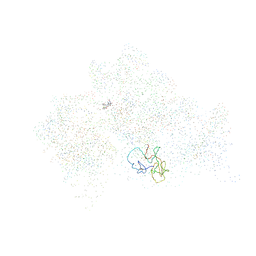 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
2FD0
 
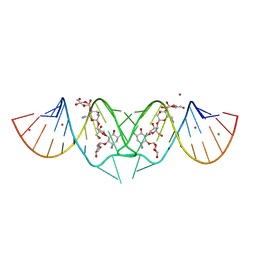 | | HIV-1 DIS kissing-loop in complex with lividomycin | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
2FSK
 
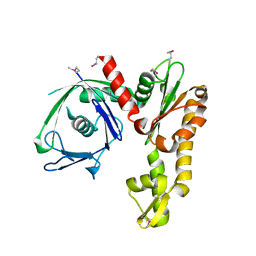 | | Crystal structure of Ta0583, an archaeal actin homolog, SeMet data | | Descriptor: | hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
1HNZ
 
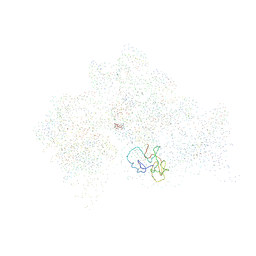 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
5X4J
 
 | | The crystal structure of Pyrococcus furiosus RecJ (Zn-soaking) | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4K
 
 | | The complex crystal structure of Pyrococcus furiosus RecJ and CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
2FSN
 
 | | Crystal structure of Ta0583, an archaeal actin homolog, complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
2L7E
 
 | |
8RH3
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to Gemcitabine | | Descriptor: | (2~{R},3~{R})-4,4-bis(fluoranyl)-2-(hydroxymethyl)oxolan-3-ol, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
4YOT
 
 | |
2LMS
 
 | | A single GalNAc residue on Threonine-106 modifies the dynamics and the structure of Interferon alpha-2a around the glycosylation site | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Interferon alpha-2 | | Authors: | Ghasriani, H, Belcourt, P.J.F, Sauve, S, Hodgson, D.J, Gingras, G, Brochu, D, Gilbert, M, Aubin, Y. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A single N-acetylgalactosamine residue at threonine 106 modifies the dynamics and structure of interferon alpha2a around the glycosylation site.
J.Biol.Chem., 288, 2013
|
|
6CFM
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a propynyl-pteridinone inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7,8-dimethyl-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a propynyl-pteridinone inhibitor
To Be Published
|
|
2KZT
 
 | | Structure of the Tandem MA-3 Region of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Waters, L.C, Strong, S.L, Oka, O, Muskett, F.W, Veverka, V, Banerjee, S, Schmedt, T, Henry, A.J, Klempnauer, K.H, Carr, M.D. | | Deposit date: | 2010-06-24 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor
J.Biol.Chem., 286, 2011
|
|
1HR1
 
 | |
