2HCR
 
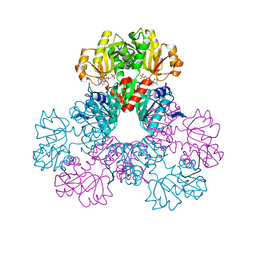 | | crystal structure of human phosphoribosyl pyrophosphate synthetase 1 in complex with AMP(ATP), cadmium and sulfate ion | | Descriptor: | ADENOSINE MONOPHOSPHATE, CADMIUM ION, Ribose-phosphate pyrophosphokinase I, ... | | Authors: | Li, S, Peng, B, Ding, J. | | Deposit date: | 2006-06-18 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site
Biochem.J., 401, 2007
|
|
3LKM
 
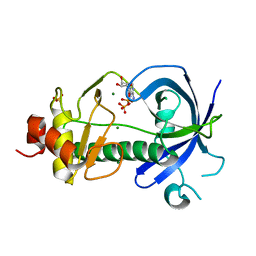 | |
7OOE
 
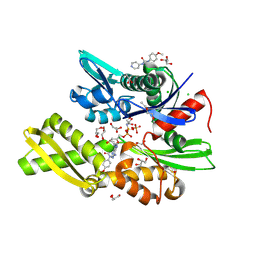 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z321318226 | | Descriptor: | AMP PHOSPHORAMIDATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.369 Å) | | Cite: | Structures of the Plasmodium falciparum heat-shock protein 70-x ATPase domain in complex with chemical fragments identify conserved and unique binding sites.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5DW8
 
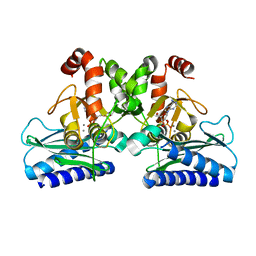 | |
7TO3
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TQD
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
5LDO
 
 | | Crystal structure of E.coli LigT complexed with 3'-AMP | | Descriptor: | RNA 2',3'-cyclic phosphodiesterase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2016-06-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase.
PLoS ONE, 12, 2017
|
|
7ZXB
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'dAMP-2'F,2'dAMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},10~{S},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-18-fluoranyl-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'dAMP-2'F,2'dAMP)
To Be Published
|
|
7ZV0
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'F,2'dAMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-13 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'F,2'dAMP)
To Be Published
|
|
7V8J
 
 | |
1OBT
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
2HBL
 
 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn, Zn, and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Exosome complex exonuclease RRP6, MANGANESE (II) ION, ... | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4J75
 
 | | Crystal Structure of a parasite tRNA synthetase, product-bound | | Descriptor: | GLYCEROL, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Koh, C.Y, Kim, J.E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Plasmodium falciparum cytosolic tryptophanyl-tRNA synthetase and its potential as a target for structure-guided drug design.
Mol.Biochem.Parasitol., 189, 2013
|
|
8G0V
 
 | |
8G0R
 
 | |
7UQ2
 
 | | Vs.4 from T4 phage in complex with cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, Vs.4 | | Authors: | Jenson, J.M, Chen, Z.J. | | Deposit date: | 2022-04-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence.
Nature, 616, 2023
|
|
8G0U
 
 | |
8G0T
 
 | |
8G0S
 
 | |
8ZB5
 
 | | Crystal structure of NudC from Mycobacterium abscessus in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
3OCW
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 3'-AMP | | Descriptor: | Lipoprotein E, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
6P63
 
 | |
6LTC
 
 | |
6LTD
 
 | |
6DNK
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
