2YOL
 
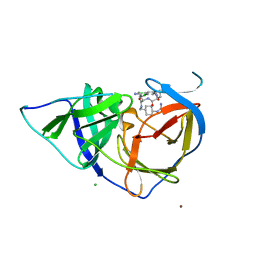 | | West Nile Virus NS2B-NS3 protease in complex with 3,4- dichlorophenylacetyl-Lys-Lys-GCMA | | Descriptor: | (S)-6-amino-N-((S)-6-amino-1-(((1r,4S)-4-guanidinocyclohexyl)methylamino)-1-oxohexan-2-yl)-2-(2-(3,4-dichlorophenyl)acetamido)hexanamide, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Hammamy, M.Z, Haase, C, Hammami, M, Hilgenfeld, R, Steinmetzer, T. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Development and Characterization of New Peptidomimetic Inhibitors of the West Nile Virus Ns2B-Ns3 Protease.
Chemmedchem, 8, 2013
|
|
2ZC5
 
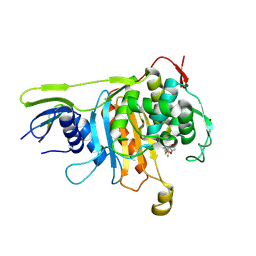 | | Penicillin-binding protein 1A (PBP 1A) acyl-enzyme complex (biapenem) from Streptococcus pneumoniae | | Descriptor: | (4R,5S)-3-(6,7-dihydro-5H-pyrazolo[1,2-a][1,2,4]triazol-4-ium-6-ylsulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-meth yl-4,5-dihydro-1H-pyrrole-2-carboxylate, Penicillin-binding protein 1A, ZINC ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Biapenem and Tebipenem Complexed with Penicillin-Binding Proteins 2X and 1A from Streptococcus pneumoniae
Antimicrob.Agents Chemother., 52, 2008
|
|
1MSK
 
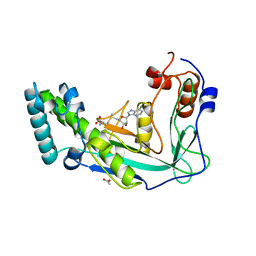 | | METHIONINE SYNTHASE (ACTIVATION DOMAIN) | | Descriptor: | ACETATE ION, COBALAMIN-DEPENDENT METHIONINE SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Dixon, M.M, Huang, S, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1996-08-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12.
Structure, 4, 1996
|
|
1N7B
 
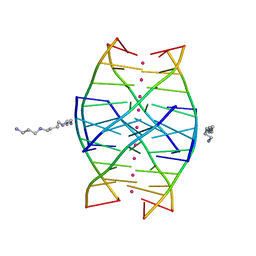 | | RIP-Radiation-damage Induced Phasing | | Descriptor: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | Authors: | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | Deposit date: | 2002-11-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1SLQ
 
 | | Crystal structure of the trimeric state of the rhesus rotavirus VP4 membrane interaction domain, VP5CT | | Descriptor: | VP4 | | Authors: | Dormitzer, P.R, Nason, E.B, Prasad, B.V.V, Harrison, S.C. | | Deposit date: | 2004-03-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural rearrangements in the membrane penetration protein of a non-enveloped virus.
Nature, 430, 2004
|
|
1NSA
 
 | |
1N7A
 
 | | RIP-Radiation-damage Induced Phasing | | Descriptor: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | Authors: | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | Deposit date: | 2002-11-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1TB6
 
 | | 2.5A Crystal Structure of the Antithrombin-Thrombin-Heparin Ternary Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Johnson, D.J, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1GPZ
 
 | | THE CRYSTAL STRUCTURE OF THE ZYMOGEN CATALYTIC DOMAIN OF COMPLEMENT PROTEASE C1R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1R COMPONENT, ... | | Authors: | Budayova-Spano, M, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2001-11-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of the Zymogen Catalytic Domain of Complement Protease C1R Reveals that a Disruptive Mechanical Stress is Required to Trigger Activation of the C1 Complex.
Embo J., 21, 2002
|
|
1JMO
 
 | | Crystal Structure of the Heparin Cofactor II-S195A Thrombin Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Baglin, T.P, Carrell, R.W, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2001-07-19 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JOU
 
 | |
2AS9
 
 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
2BP4
 
 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide in TFE-water (80-20) solution | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Changes of Region 1-16 of the Alzheimer Disease Amyloid Beta-Peptide Upon Zinc Binding and in Vitro Aging.
J.Biol.Chem., 281, 2006
|
|
1YU6
 
 | | Crystal Structure of the Subtilisin Carlsberg:OMTKY3 Complex | | Descriptor: | CALCIUM ION, Ovomucoid, Subtilisin Carlsberg | | Authors: | Maynes, J.T, Cherney, M.M, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the subtilisin Carlsberg-OMTKY3 complex reveals two different ovomucoid conformations.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2D91
 
 | | Structure of HYPER-VIL-lysozyme | | Descriptor: | IODIDE ION, Lysozyme C | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D97
 
 | | Structure of VIL-xylanase | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1M7G
 
 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1M7H
 
 | | Crystal Structure of APS kinase from Penicillium Chrysogenum: Structure with APS soaked out of one dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Adenylylsulfate kinase, ... | | Authors: | Lansdon, E.B, Sege, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1OB2
 
 | | E. coli elongation factor EF-Tu complexed with the antibiotic kirromycin, a GTP analog, and Phe-tRNA | | Descriptor: | ELONGATION FACTOR TU, KIRROMYCIN, MAGNESIUM ION, ... | | Authors: | Kristensen, O, Nissen, P, Nyborg, J. | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Kirromycin Defines a Specific Domain Arrangement of Elongation Factor EF-TU
To be Published
|
|
1DM4
 
 | | SER195ALA MUTANT OF HUMAN THROMBIN COMPLEXED WITH FIBRINOPEPTIDE A (7-16) | | Descriptor: | PROTEIN (ALPHA THROMBIN:LIGHT CHAIN), PROTEIN (FIBRINOPEPTIDE), PROTEIN (MUTANT ALPHA THROMBIN:HEAVY CHAIN) | | Authors: | Krishnan, R, Sadler, E.J, Tulinsky, A. | | Deposit date: | 1999-12-13 | | Release date: | 2000-01-19 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ser195Ala mutant of human alpha--thrombin complexed with fibrinopeptide A(7--16): evidence for residual catalytic activity.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1BML
 
 | |
1QXQ
 
 | |
1QX9
 
 | |
6C6W
 
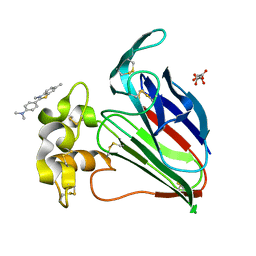 | | Structure of a Complex Between Thaumatin and Thioflavin T | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | McPherson, A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-07 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Investigation into the binding of dyes within protein crystals.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E0D
 
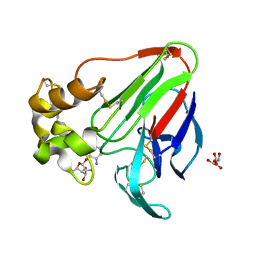 | | X-ray structure of a complex of thaumatin with xylene cyanol | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | McPherson, A. | | Deposit date: | 2018-07-06 | | Release date: | 2018-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Investigation into the binding of dyes within protein crystals.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
