8CO2
 
 | |
8C3L
 
 | |
1TQ0
 
 | | Crystal structure of the potent anticoagulant thrombin mutant W215A/E217A in free form | | Descriptor: | Prothrombin | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
2BRH
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | N-(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)GLYCINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
1G74
 
 | |
3VQD
 
 | | HIV-1 IN core domain in complex with 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid | | Descriptor: | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQQ
 
 | | HIV-1 integrase core domain in complex with 2,1,3-benzothiadiazol-4-amine | | Descriptor: | 2,1,3-benzothiadiazol-4-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQ9
 
 | | HIV-1 IN core domain in complex with 6-fluoro-1,3-benzothiazol-2-amine | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, POL polyprotein | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQP
 
 | | HIV-1 IN core domain in complex with 2,3-dihydro-1,4-benzodioxin-5-ylmethanol | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-5-ylmethanol, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4L9G
 
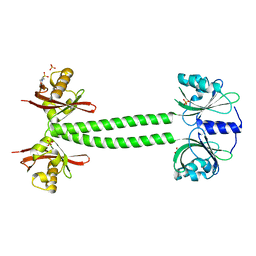 | | Structure of PpsR N-Q-PAS1 from Rb. sphaeroides | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Heintz, U, Meinhart, A, Schlichting, I, Winkler, A. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multi-PAS domain-mediated protein oligomerization of PpsR from Rhodobacter sphaeroides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1TVP
 
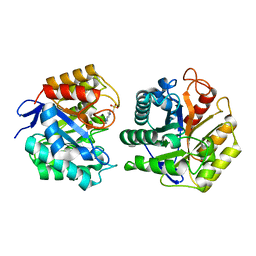 | |
7KZH
 
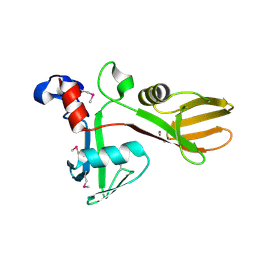 | |
1TTZ
 
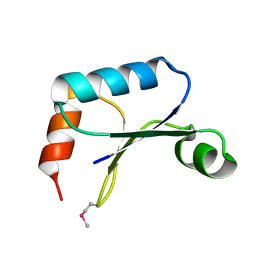 | | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Ho, C.K, Cooper, B, Ma, L.-C, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris
To be Published
|
|
7KPY
 
 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
1G7N
 
 | |
1TIY
 
 | | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160 | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Kuzin, A.P, Vorobiev, S, Edstrom, W, Forouhar, F, Acton, T, Shastry, R, Ma, L.-C, Chiang, Y.-W, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS
NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160
To be published
|
|
6CBF
 
 | |
1T22
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9, orthorhombic crystal | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
2Y7P
 
 | | DntR Inducer Binding Domain in Complex with Salicylate. Trigonal crystal form | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-HYDROXYBENZOIC ACID, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Devesse, L, Smirnova, I, Lonneborg, R, Kapp, U, Brzezinski, P, Leonard, G.A, Dian, C. | | Deposit date: | 2011-02-01 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Dntr Inducer Binding Domains in Complex with Salicylate Offer Insights Into the Activation of Lysr-Type Transcriptional Regulators.
Mol.Microbiol., 81, 2011
|
|
2Y5L
 
 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7KL3
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D bound to RNA oligomer AG*CAUC (*uncleaveable bond, -UC disordered) | | Descriptor: | 5'-O-sulfocytidine, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cuypers, M.G, Kumar, G, White, S.W. | | Deposit date: | 2020-10-28 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
1T8M
 
 | | CRYSTAL STRUCTURE OF THE P1 HIS BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-13 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
1IPB
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GPPPA | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1T1X
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-4L | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1T7C
 
 | | CRYSTAL STRUCTURE OF THE P1 GLU BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-09 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
