6OF1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with dirithromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khabibullina, N.F, Tereshchenkov, A.G, Komarova, E.S, Syroegin, E.A, Shiriaev, D.I, Paleskava, A, Kartsev, V.G, Bogdanov, A.A, Konevega, A.L, Dontsova, O.A, Sergiev, P.V, Osterman, I.A, Polikanov, Y.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Dirithromycin Bound to the Bacterial Ribosome Suggests New Ways for Rational Improvement of Macrolides.
Antimicrob.Agents Chemother., 63, 2019
|
|
8ON7
 
 | | FMRFa-bound Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ONA
 
 | | FMRFa-bound Malacoceros FaNaC1 in lipid nanodiscs in presence of diminazene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide, neuropeptide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ON8
 
 | | Apo Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated sodium channel 1 | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ON9
 
 | | ASSFVRIa-bound Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASSFVRIamide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4ZSN
 
 | |
5LQ5
 
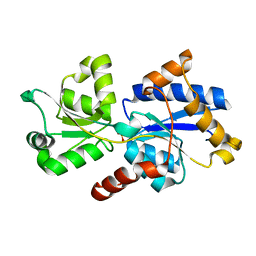 | | 1.46 A resolution structure of PhnD1 from Prochlorococcus marinus (MIT 9301) in complex with phosphite | | Descriptor: | PHOSPHITE ION, Putative phosphonate binding protein for ABC transporter | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-08-16 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
7AB8
 
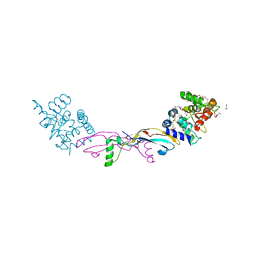 | | Crystal structure of a GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
4V30
 
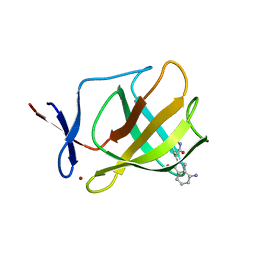 | |
4V31
 
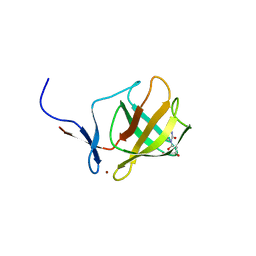 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, CEREBLON ISOFORM 4, CITRATE ANION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
4V32
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Y101F mutant | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
4V2Y
 
 | |
4V2Z
 
 | |
6T9T
 
 | | Matriptase in complex with the synthetic inhibitor (S)-3-(3-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-2-((3'-fluoro-4'-(hydroxymethyl)-[1,1'-biphenyl])-3-sulfonamido)-3-oxopropyl)benzimidamide (MI-1904) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-[3-fluoranyl-4-(hydroxymethyl)phenyl]phenyl]sulfonylamino ]propanoyl]piperidin-4-yl]urea, CHLORIDE ION, Suppressor of tumorigenicity 14 protein | | Authors: | Mueller, J.M, Merkl, S, Keils, A, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
2VO4
 
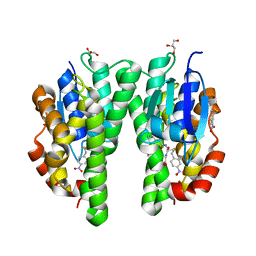 | | Glutathione transferase from Glycine max | | Descriptor: | 2,4-D INDUCIBLE GLUTATHIONE S-TRANSFERASE, 4-NITROPHENYL METHANETHIOL, GLYCEROL, ... | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and Functional Characterization of the Fluorodifen-Inducible Glutathione Transferase from Glycine Max Reveals an Active Site Topography Suited for Diphenylether Herbicides and a Novel L-Site.
J.Mol.Biol., 385, 2009
|
|
6U42
 
 | | Natively decorated ciliary doublet microtubule | | Descriptor: | DC1, DC2, DC3, ... | | Authors: | Ma, M, Stoyanova, M, Rademacher, G, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Decorated Ciliary Doublet Microtubule.
Cell, 179, 2019
|
|
5J3C
 
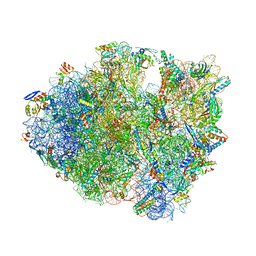 | |
8KH5
 
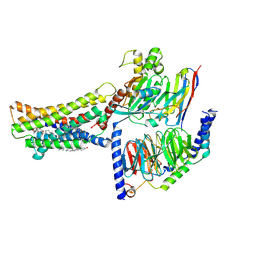 | | Cryo-EM structure of the GPR174-Gs complex bound to endogenous lysoPS | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nie, Y, Qiu, Z, Zheng, S, Chen, S. | | Deposit date: | 2023-08-21 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
8KGK
 
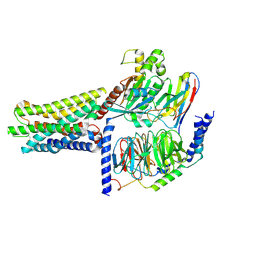 | | Cryo-EM structure of the GPR61-Gs complex | | Descriptor: | G-protein coupled receptor 61, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nie, Y, Qiu, Z, Zheng, S. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
8KH4
 
 | | Cryo-EM structure of the GPR161-Gs complex | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nie, Y, Qiu, Z, Zheng, S, Chen, S. | | Deposit date: | 2023-08-21 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive G s signaling.
Nat Commun, 14, 2023
|
|
5J4B
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (co-crystallized) and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
5I4L
 
 | | Crystal structure of Amicoumacin A bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I.V, Yusupova, G, Yusupov, M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Amicoumacin A induces cancer cell death by targeting the eukaryotic ribosome.
Sci Rep, 6, 2016
|
|
5J30
 
 | |
5J4C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (soaked) and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
5J4D
 
 | |
