9FZP
 
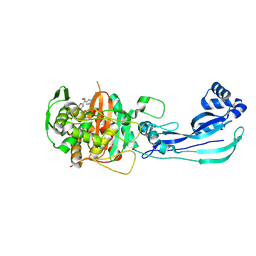 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with cefepime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZ8
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZO
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Peptidoglycan D,D-transpeptidase FtsI, SULFATE ION | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
9FZE
 
 | | Pseudomonas aeruginosa penicillin binding protein 3 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Smith, H.G, Allen, M.D, Basak, S, Aniebok, V, Beech, M.J, Alshref, F.M, Farley, A.J.M, Schofield, C.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Pseudomonas aeruginosa penicillin binding protein 3 inhibition by the siderophore-antibiotic cefiderocol.
Chem Sci, 15, 2024
|
|
5AN5
 
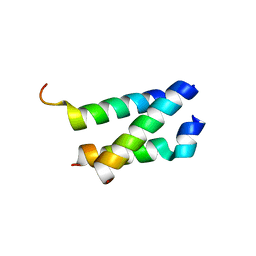 | | B. subtilis GpsB C-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, GLYCEROL | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
7RCZ
 
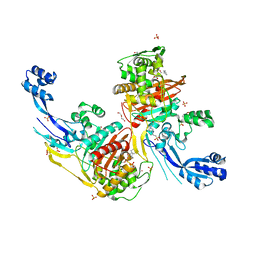 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7ZG8
 
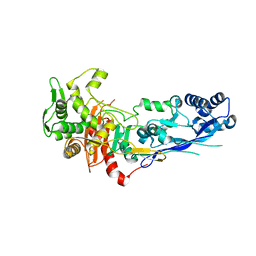 | |
8VBW
 
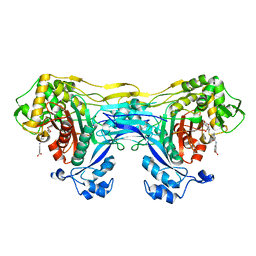 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBU
 
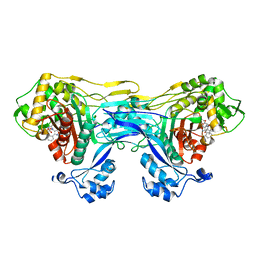 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
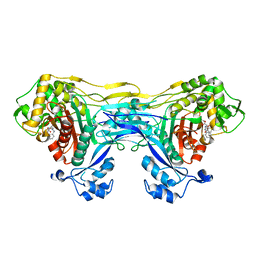 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
1HVB
 
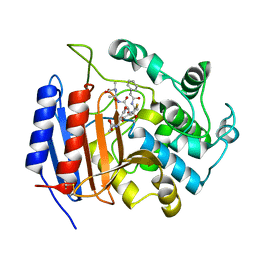 | | CRYSTAL STRUCTURE OF STREPTOMYCES R61 DD-PEPTIDASE COMPLEXED WITH A NOVEL CEPHALOSPORIN ANALOG OF CELL WALL PEPTIDOGLYCAN | | Descriptor: | 5-{3-(S)-(4-(R)-ACETYLAMINO-4-CARBOXY-BUTYRYLAMINO)-3-[1-(R)-(1-(R)-CARBOXY-ETHYLCARBAMOYL)-ETHYLCARBAMOYL]-PROPYL}-2-( CARBOXY-PHENYLACETYLAMINO-METHYL)-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | | Authors: | McDonough, M.A, Lee, W, Silvaggi, N.R, Mobashery, S, Kelly, J.A. | | Deposit date: | 2001-01-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A 1.2-A snapshot of the final step of bacterial cell wall biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7KCX
 
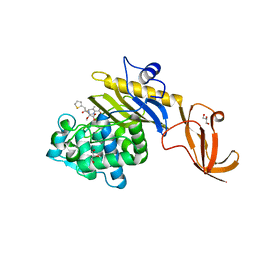 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCY
 
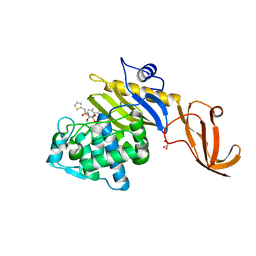 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCW
 
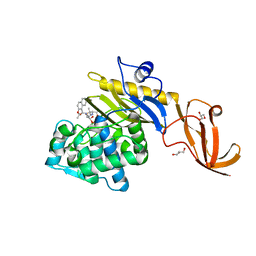 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A, Strynadka, N.C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
5E31
 
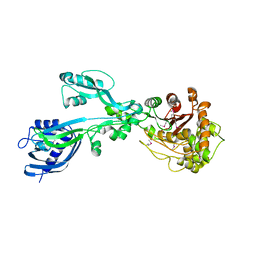 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
3VMA
 
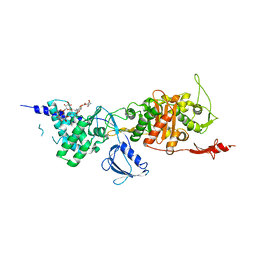 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Huang, C.Y, Sung, M.T, Lai, Y.T, Chou, L.Y, Shih, H.W, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FWL
 
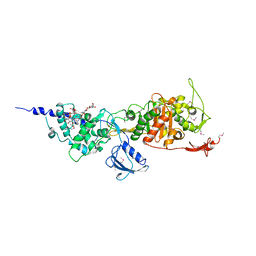 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Sung, M.T, Lai, Y.T, Huang, C.Y, Chou, L.Y, Wong, C.H, Ma, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5DVY
 
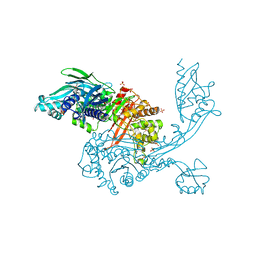 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
4CJN
 
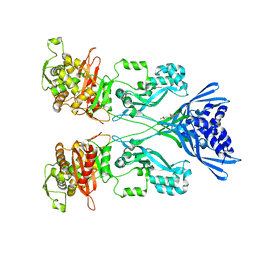 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | Descriptor: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2013-12-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
3PBR
 
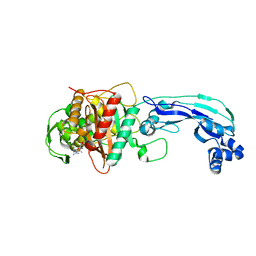 | | Crystal structure of PBP3 complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBO
 
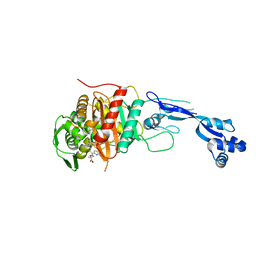 | | Crystal structure of PBP3 complexed with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBT
 
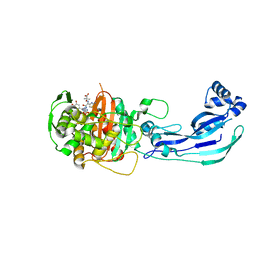 | | Crystal structure of PBP3 complexed with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 3 | | Authors: | Han, S, Evdokimov, A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBS
 
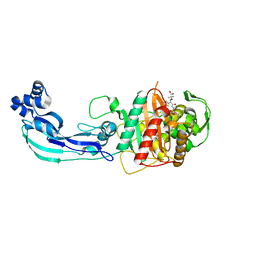 | | Crystal structure of PBP3 complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBQ
 
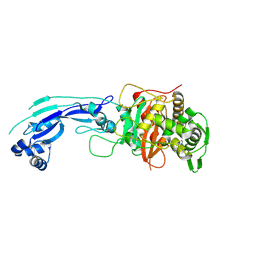 | | Crystal structure of PBP3 complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2J5U
 
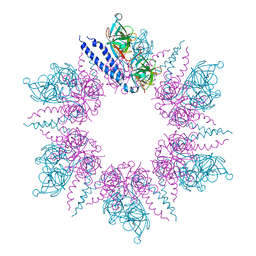 | | MreC Lysteria monocytogenes | | Descriptor: | MREC PROTEIN | | Authors: | van den Ent, F, Leaver, M, Bendezu, F, Errington, J, de Boer, P, Lowe, J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric Structure of the Cell Shape Protein Mrec and its Functional Implications.
Mol.Microbiol., 62, 2006
|
|
