8DQ4
 
 | |
8DQ3
 
 | |
2C83
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 | | Descriptor: | HYPOTHETICAL PROTEIN PM0188 | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
8BVO
 
 | |
8BXR
 
 | |
8BW6
 
 | | Titin FnIII-domain I110 (I/A6) from the MIR region | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mayans, O, Fleming, J.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Immunological and Structural Characterization of Titin Main Immunogenic Region; I110 Domain Is the Target of Titin Antibodies in Myasthenia Gravis.
Biomedicines, 11, 2023
|
|
2NZU
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2USN
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-141803 | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION, ... | | Authors: | Finzel, B.C, Bryant Junior, G.L, Baldwin, E.T. | | Deposit date: | 1998-06-09 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity.
Protein Sci., 7, 1998
|
|
1XHO
 
 | | Chorismate mutase from Clostridium thermocellum Cth-682 | | Descriptor: | Chorismate mutase, UNKNOWN ATOM OR ION | | Authors: | Xu, H, Chen, L, Lee, D, Habel, J.E, Nguyen, J, Chang, S.-H, Kataeva, I, Chang, J, Zhao, M, Yang, H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Away from the edge II: in-house Se-SAS phasing with chromium radiation.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CD5
 
 | |
2CD3
 
 | |
8F4O
 
 | | Apo structure of the TPP riboswitch aptamer domain | | Descriptor: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | Authors: | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
1ZFF
 
 | | TTC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*TP*TP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZID
 
 | |
4APM
 
 | | Crystal Structure of AMA1 from Babesia divergens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, APICAL MEMBRANE ANTIGEN 1, ... | | Authors: | Tonkin, M.L, Crawford, J, Lebrun, M.L, Boulanger, M.J. | | Deposit date: | 2012-04-04 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Babesia Divergens and Neospora Caninum Apical Membrane Antigen 1 (Ama1) Structures Reveal Selectivity and Plasticity in Apicomplexan Parasite Host Cell Invasion.
Protein Sci., 22, 2013
|
|
4APL
 
 | |
2ATC
 
 | | CRYSTAL AND MOLECULAR STRUCTURES OF NATIVE AND CTP-LIGANDED ASPARTATE CARBAMOYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE, CATALYTIC CHAIN, REGULATORY CHAIN, ... | | Authors: | Honzatko, R.B, Crawford, J.L, Monaco, H.L, Ladner, J.E, Edwards, B.F.P, Evans, D.R, Warren, S.G, Wiley, D.C, Ladner, R.C, Lipscomb, W.N. | | Deposit date: | 1982-03-24 | | Release date: | 1982-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli.
J.Mol.Biol., 160, 1982
|
|
6J08
 
 | | Crystal structure of human MAJIN and TERB2 | | Descriptor: | Membrane-anchored junction protein, Telomere repeats-binding bouquet formation protein 2 | | Authors: | Wang, Y, Chen, Y, Wu, J, Huang, C, Lei, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The meiotic TERB1-TERB2-MAJIN complex tethers telomeres to the nuclear envelope.
Nat Commun, 10, 2019
|
|
1ZEY
 
 | | CGG A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*GP*CP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZF6
 
 | | TGG DUPLEX A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*AP*TP*GP*GP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4BBW
 
 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | Descriptor: | SIALIDASE (NEURAMINIDASE) | | Authors: | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-08-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
5ZDS
 
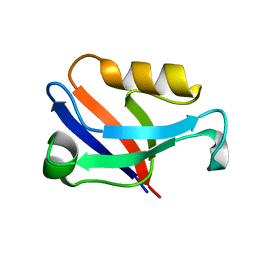 | | Crystal structure of the second PDZ domain of Frmpd2 | | Descriptor: | FERM and PDZ domain-containing 2 | | Authors: | Lu, X, Wang, X.F. | | Deposit date: | 2018-02-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The second PDZ domain of scaffold protein Frmpd2 binds to GluN2A of NMDA receptors.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
2AT1
 
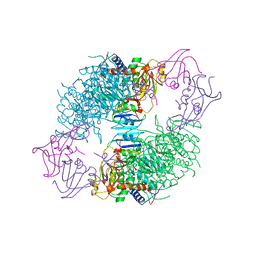 | |
4AT1
 
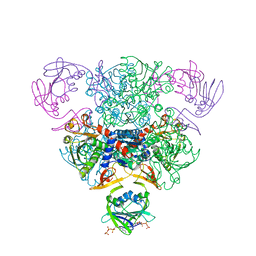 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
5ZYS
 
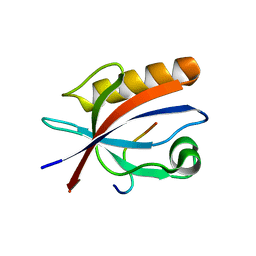 | | Structure of Nephrin/MAGI1 complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Nephrin | | Authors: | Weng, Z.F, Shng, Y, Zhu, J.W, Zhang, R.G. | | Deposit date: | 2018-05-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis of Highly Specific Interaction between Nephrin and MAGI1 in Slit Diaphragm Assembly and Signaling.
J. Am. Soc. Nephrol., 29, 2018
|
|
