6W3R
 
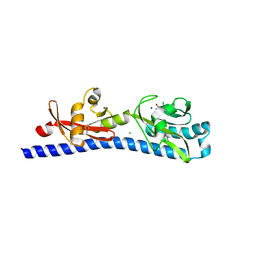 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 3-methylisoleucine | | Descriptor: | 3-methyl-L-alloisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
2WGF
 
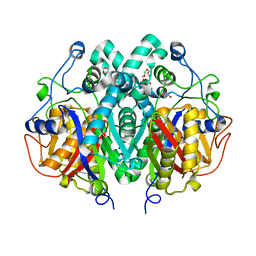 | | Crystal structure of Mycobacterium tuberculosis C171Q KasA variant | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Luckner, S.R, Kisker, C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Mycobacterium Tuberculosis Kasa Show Mode of Action within Cell Wall Biosynthesis and its Inhibition by Thiolactomycin
Structure, 17, 2009
|
|
4R3W
 
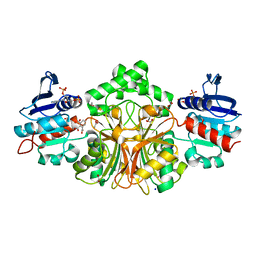 | | Crystal Structure Analysis of the 1,2,3-tricarboxylate benzoic acid bound to sp-ASADH-2'5'-ADP complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-2'-5'-DIPHOSPHATE, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QWX
 
 | | yCP in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5OAB
 
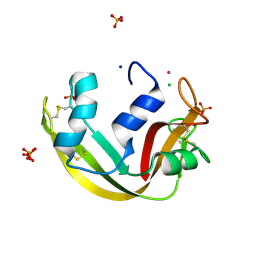 | | A novel crystal form of human RNase6 at atomic resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.111 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6VND
 
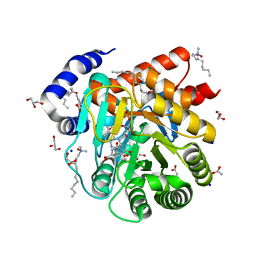 | | Quaternary Complex of human dihydroorotate dehydrogenase (DHODH) with flavin mononucleotide (FMN), orotic acid and AG-636 | | Descriptor: | 1-methyl-5-(2'-methyl[1,1'-biphenyl]-4-yl)-1H-benzotriazole-7-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Selective Vulnerability to Pyrimidine Starvation in Hematologic Malignancies Revealed by AG-636, a Novel Clinical-Stage Inhibitor of Dihydroorotate Dehydrogenase.
Mol.Cancer Ther., 19, 2020
|
|
2OZQ
 
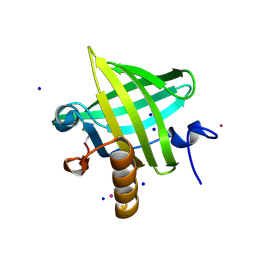 | | Crystal Structure of apo-MUP | | Descriptor: | CADMIUM ION, Novel member of the major urinary protein (Mup) gene family, SODIUM ION | | Authors: | Dennis, C.A, Homans, S.W, Phillips, S.E.V, Syme, N.R. | | Deposit date: | 2007-02-27 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origin of heat capacity changes in a "nonclassical" hydrophobic interaction.
Chembiochem, 8, 2007
|
|
6W3O
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 4-methylisoleucine | | Descriptor: | 4-methylisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
4R54
 
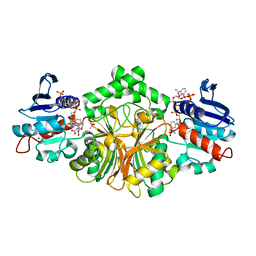 | | Complex crystal structure of sp-Aspartate-Semialdehyde-Dehydrogenase with 3-carboxy-ethyl-phthalic acid | | Descriptor: | 3-(2-carboxyethyl)benzene-1,2-dicarboxylic acid, ACETATE ION, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8GF8
 
 | | Cryo-EM structure of human TRPV1 in cNW11 nanodisc and soybean lipids | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
8GHF
 
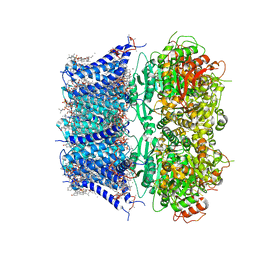 | | cryo-EM structure of hSlo1 in plasma membrane vesicles | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Tao, X, Zhao, C, MacKinnon, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Membrane protein isolation and structure determination in cell-derived membrane vesicles.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GF9
 
 | | Cryo-EM structure of human TRPV1 in cNW11 nanodisc and POPC:POPE:POPG lipids | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
6UJ7
 
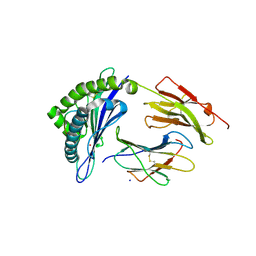 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
8GFA
 
 | | Cryo-EM structure of human TRPV1 in complex with the analgesic drug SB-366791 | | Descriptor: | (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
2OQD
 
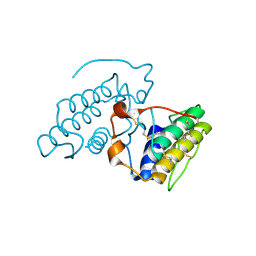 | | Crystal Structure of BthTX-II | | Descriptor: | Phospholipase A2 | | Authors: | Correa, L.C, Marchi-Salvador, D.P, Cintra, A.C.O, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of a myotoxic Asp49-phospholipase A(2) with low catalytic activity: Insights into Ca(2+)-independent catalytic mechanism.
Biochim.Biophys.Acta, 1784, 2008
|
|
3FFZ
 
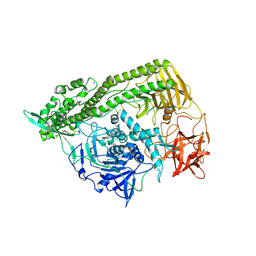 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
4PA3
 
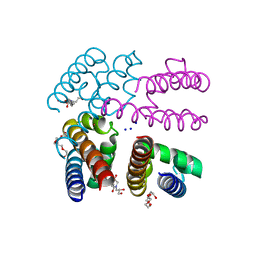 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5NZ3
 
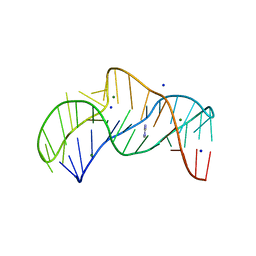 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
6W1Q
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the second illumination at 2.27 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3N7Z
 
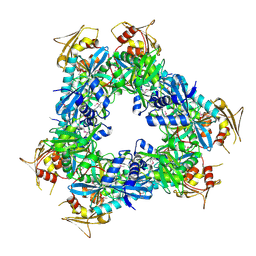 | | Crystal structure of acetyltransferase from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, SODIUM ION | | Authors: | Chang, C, Wu, R, Gornicki, P, Zhang, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and Structural Analysis of an Eis Family Aminoglycoside Acetyltransferase from Bacillus anthracis.
Biochemistry, 54, 2015
|
|
6VUT
 
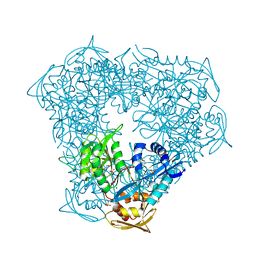 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT392 | | Descriptor: | 2-[(4-amino-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-2-yl)sulfanyl]-N-[2-(morpholin-4-yl)ethyl]acetamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
4TPU
 
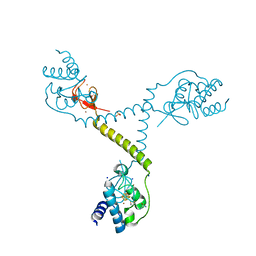 | | CRYSTAL STRUCTURE OF FERREDOXIN-DEPENDENT DISULFIDE REDUCTASE FROM METHANOSARCINA ACETIVORANS | | Descriptor: | BROMIDE ION, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A.K, Yennawar, H.P, Yennawar, N.H, Ferry, J.G. | | Deposit date: | 2014-06-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural and Biochemical Characterization of a Ferredoxin:Thioredoxin Reductase-like Enzyme from Methanosarcina acetivorans.
Biochemistry, 54, 2015
|
|
4TOF
 
 | | 1.65A resolution structure of BfrB (C89S, K96C) crystal form 1 from Pseudomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bacterioferritin, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRZ
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp13-nsp14 (C13) cut site sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
