8TV4
 
 | | NMR structure of temporin L in solution | | Descriptor: | Temporin-1Tl peptide | | Authors: | McShan, A.C, Jia, R, Halim, M.A. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiviral peptides inhibiting the main protease of SARS-CoV-2 investigated by computational screening and in vitro protease assay.
J.Pept.Sci., 30, 2024
|
|
1FQZ
 
 | | NMR VALIDATED MODEL OF DOMAIN IIID OF HEPATITIS C VIRUS INTERNAL RIBOSOME ENTRY SITE | | Descriptor: | HEPATITIS C VIRUS IRES DOMAIN IIID | | Authors: | Klinck, R, Westhof, E, Walker, S, Afshar, M, Collier, A, Aboul-ela, F. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A potential RNA drug target in the hepatitis C virus internal ribosomal entry site.
RNA, 6, 2000
|
|
1BO0
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN-3, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (MONOCYTE CHEMOATTRACTANT PROTEIN-3) | | Authors: | Kwon, D, Lee, D, Sykes, B.D, Kim, K.-S. | | Deposit date: | 1998-08-10 | | Release date: | 1999-10-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a monomeric chemokine: monocyte chemoattractant protein-3.
FEBS Lett., 395, 1996
|
|
8X1V
 
 | |
3F2E
 
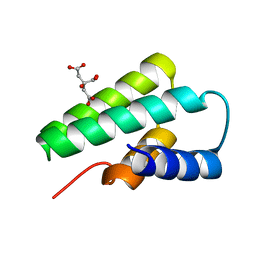 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
6N68
 
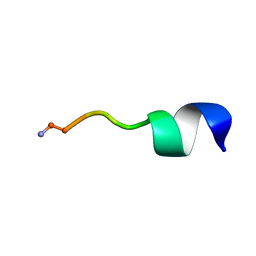 | |
1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DNG
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL ACIDIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DN3
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | HUMAN PLATELET FACTOR 4, SEGMENT 59-73 | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-12 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1GO1
 
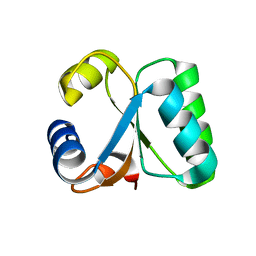 | |
6O3S
 
 | | NMR solution structure of Luffin P1 | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Rosengren, K.J, Payne, C. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Ancient Peptide Family Buried within Vicilin Precursors.
Acs Chem.Biol., 14, 2019
|
|
5NAM
 
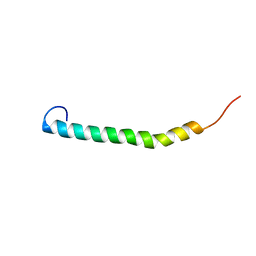 | |
5N5C
 
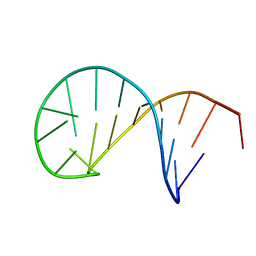 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
5NAO
 
 | |
5NWU
 
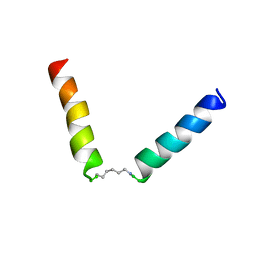 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | wtFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWV
 
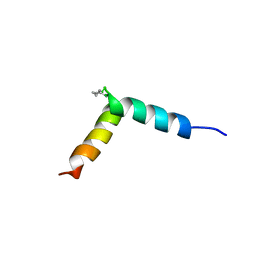 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWW
 
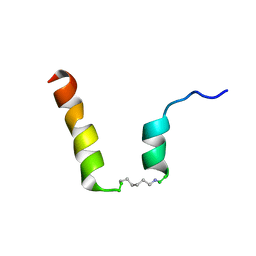 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NVP
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
1QWA
 
 | | NMR structure of 5'-r(GGAUGCCUCCCGAGUGCAUCC): an RNA hairpin derived from the mouse 5'ETS that binds nucleolin RBD12. | | Descriptor: | 18S ribosomal RNA, 5'ETS | | Authors: | Finger, L.D, Trantirek, L, Johansson, C, Feigon, J. | | Deposit date: | 2003-09-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of Stem-loop RNAs that Bind to the Two N-terminal RNA Binding Domains of Nucleolin
Nucleic Acids Res., 31, 2003
|
|
6QXC
 
 | | NMR structure of peptide 8, characterized by a trans-4-cyclohexyl-Pro, with a dramatic reduction in activity on E. coli ATCC and lost effect on P. aeruginosa. | | Descriptor: | PHE-VAL-TCP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
1T2Y
 
 | | NMR solution structure of the protein part of Cu6-Neurospora crassa MT | | Descriptor: | Metallothionein | | Authors: | Cobine, P.A, McKay, R.T, Zangger, K, Dameron, C.T, Armitage, I.M. | | Deposit date: | 2004-04-23 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Cu metallothionein from the fungus Neurospora crassa
Eur.J.Biochem., 271, 2004
|
|
1ILF
 
 | |
7P27
 
 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
2RQM
 
 | | NMR Solution Structure of Mesoderm Development (MESD) - open conformation | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Lighthouse, J.K, Werther, T, Andersen, O.M, Diehl, A, Schmieder, P, Holdener, B.C, Oschkinat, H. | | Deposit date: | 2009-08-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding
Structure, 19, 2011
|
|
2RS4
 
 | | NMR structure of stereo-array isotope labelled (SAIL) peptidyl-prolyl cis-trans isomerase from E. coli (EPPIb) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase B | | Authors: | Takeda, M, Jee, J, Ono, A.M, Okuma, K, Terauchi, T, Kainosho, M. | | Deposit date: | 2011-07-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hydrogen exchange study on the hydroxyl groups of serine and threonine residues in proteins and structure refinement using NOE restraints with polar side-chain groups
J.Am.Chem.Soc., 133, 2011
|
|
