4Y2W
 
 | |
5L3S
 
 | | Structure of the GTPase heterodimer of crenarchaeal SRP54 and FtsY | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
8BBQ
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, GLYCEROL, ... | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
8BBR
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, SULFATE ION | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
5KYH
 
 | | Structure of Iho670 Flagellar-like Filament | | Descriptor: | Iho670 | | Authors: | Braun, T, Vos, M, Kalisman, N, Sherman, N.E, Rachel, R, Wirth, R, Schroeder, G.F, Egelman, E.H. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
6O9Y
 
 | | Structure of human PARG complexed with JA2-8 | | Descriptor: | 7-[(2S)-2-hydroxy-3-(morpholin-4-yl)propyl]-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6OAL
 
 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Ahmed, Z, Warden, S, Tainer, J.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
4XZR
 
 | |
8IFE
 
 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
6OA3
 
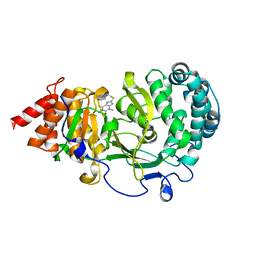 | | Structure of human PARG complexed with JA2131 | | Descriptor: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
5LOZ
 
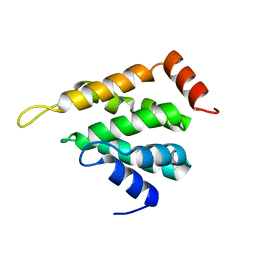 | | STRUCTURE OF YEAST ENT1 ENTH DOMAIN | | Descriptor: | Epsin-1 | | Authors: | Tanner, N, Prag, G. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A bacterial genetic selection system for ubiquitylation cascade discovery.
Nat.Methods, 13, 2016
|
|
5LL6
 
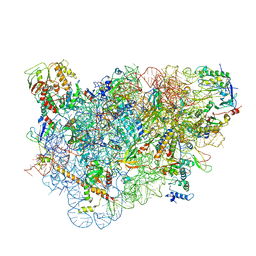 | | Structure of the 40S ABCE1 post-splitting complex in ribosome recycling and translation initiation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Heuer, A, Gerovac, M, Schmidt, C, Trowitzsch, S, Preis, A, Koetter, P, Berninghausen, O, Becker, T, Beckmann, R, Tampe, R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5JQ4
 
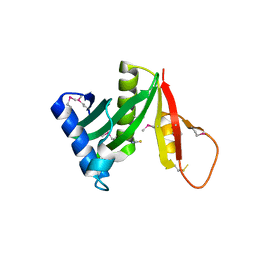 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase SACOL1063, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
7UXG
 
 | | Crystal structure of putative serine protease YdgD from Escherichia coli | | Descriptor: | Serine protease | | Authors: | Stogios, P.J, Michalska, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of putative serine protease YdgD from Escherichia coli
To Be Published
|
|
7V09
 
 | | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein | | Descriptor: | MAGNESIUM ION, Multiple sugar transport system periplasmic sugar-binding protein | | Authors: | Stogios, P.J, Skarina, T, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein
To Be Published
|
|
6NW8
 
 | | SOLUTION STRUCTURE OF CN29, A TOXIN FROM CENTRUROIDES NOXIUS SCORPION VENOM | | Descriptor: | Cn29 | | Authors: | Delepierre, M, Gurrola, G.B, Possani, L.D, Guijarro, J.I. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Cn29, a novel orphan peptide found in the venom of the scorpion Centruroides noxius: Structure and function.
Toxicon, 167, 2019
|
|
6OAK
 
 | | Structure of human PARG complexed with JA2131 | | Descriptor: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Ahmed, Z, Warden, S, Tainer, J.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7UFG
 
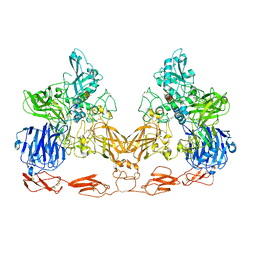 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
6OA0
 
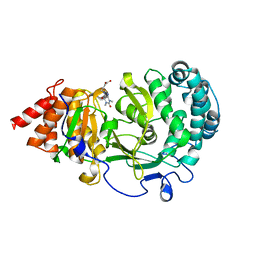 | | Structure of human PARG complexed with JA2-9 | | Descriptor: | 4-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)butanoic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUO
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA H135A mutant, complex with tobramycin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, COENZYME A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UUM
 
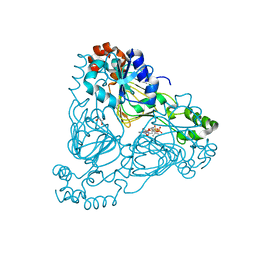 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with paromomycin and coenzyme A | | Descriptor: | Aminocyclitol acetyltransferase ApmA, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7USL
 
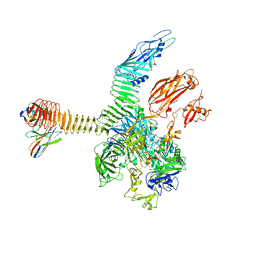 | | Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
