4GBD
 
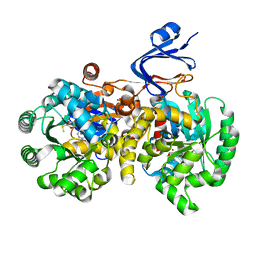 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
1TLV
 
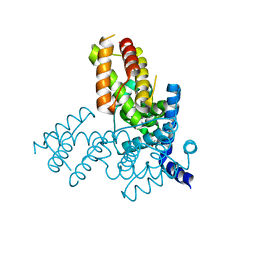 | | Structure of the native and inactive LicT PRD from B. subtilis | | Descriptor: | Transcription antiterminator licT | | Authors: | Graille, M, Zhou, C.-Z, Receveur-Brechot, V, Collinet, B, Declerck, N, van Tilbeurgh, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of the LicT Transcriptional Antiterminator Involves a Domain Swing/Lock Mechanism Provoking Massive Structural Changes
J.Biol.Chem., 280, 2005
|
|
4G8K
 
 | |
4GLK
 
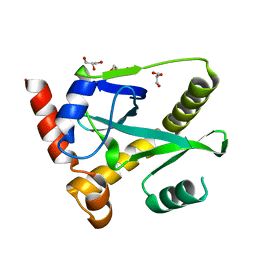 | | Structure and activity of AbiQ, a lactococcal anti-phage endoribonuclease belonging to the type-III toxin-antitoxin system | | Descriptor: | AbiQ, GLYCEROL | | Authors: | Samson, J, Spinelli, S, Cambillau, C, Moineau, S. | | Deposit date: | 2012-08-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system.
Mol.Microbiol., 87, 2013
|
|
4LT9
 
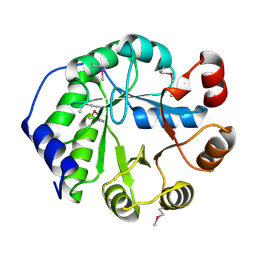 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
1TUV
 
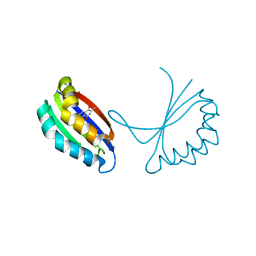 | | Crystal structure of YgiN in complex with menadione | | Descriptor: | MENADIONE, Protein ygiN | | Authors: | Adams, M.A, Jia, Z. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Evidence for an Enzymatic Quinone Redox Cycle in Escherichia coli: IDENTIFICATION OF A NOVEL QUINOL MONOOXYGENASE
J.Biol.Chem., 280, 2005
|
|
3K1X
 
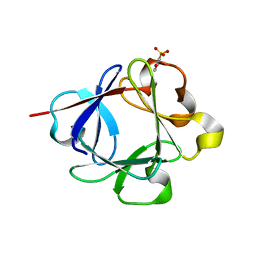 | | Acidic Fibroblast Growth Factor (FGF-1) complexed with dobesilate | | Descriptor: | 2,5-dihydroxybenzenesulfonic acid, Heparin-binding growth factor 1 | | Authors: | Romero, A, Fernandez, I.S, Gimenez-Gallego, G. | | Deposit date: | 2009-09-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Gentisic acid, a compound associated with plant defense and a metabolite of aspirin, heads a new class of in vivo fibroblast growth factor inhibitors.
J.Biol.Chem., 285, 2010
|
|
4HU8
 
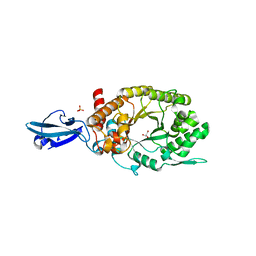 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
3O6U
 
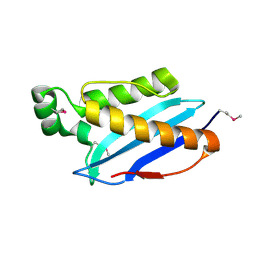 | | Crystal Structure of CPE2226 protein from Clostridium perfringens. Northeast Structural Genomics Consortium Target CpR195 | | Descriptor: | uncharacterized protein CPE2226 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CPE2226 protein from Clostridium perfringens.
To be Published
|
|
3O7U
 
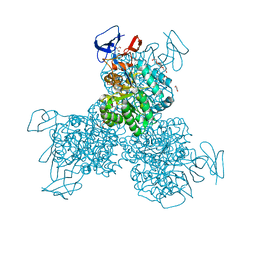 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and phosphono-cytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Three-dimensional structure and catalytic mechanism of Cytosine deaminase.
Biochemistry, 50, 2011
|
|
3P3B
 
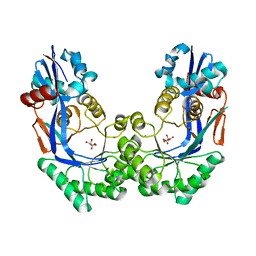 | | CRYSTAL STRUCTURE OF Galacturonate DEHYDRATASE FROM GEOBACILLUS SP. COMPLEXED WITH D-TARTRATE | | Descriptor: | D(-)-TARTARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | CRYSTAL STRUCTURE OF Galacturonate DEHYDRATASE FROM GEOBACILLUS SP. COMPLEXED WITH D-TARTRATE.
To be Published
|
|
3PKN
 
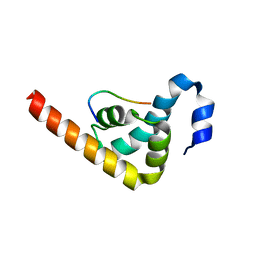 | | Crystal structure of MLLE domain of poly(A) binding protein in complex with PAM2 motif of La-related protein 4 (LARP4) | | Descriptor: | IODIDE ION, La-related protein 4, Polyadenylate-binding protein 1, ... | | Authors: | Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | La-Related Protein 4 Binds Poly(A), Interacts with the Poly(A)-Binding Protein MLLE Domain via a Variant PAM2w Motif, and Can Promote mRNA Stability.
Mol.Cell.Biol., 31, 2011
|
|
3PJV
 
 | |
3PU2
 
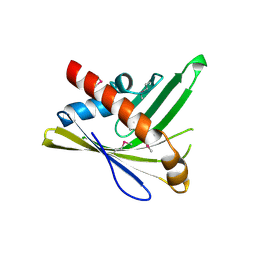 | | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides. Northeast Structural Genomics Consortium Target RhR263. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides.
To be Published
|
|
3PNZ
 
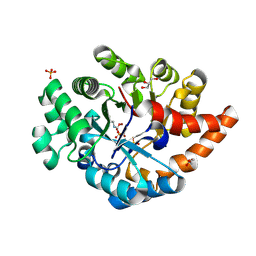 | | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphotriesterase family protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-11-20 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.5983 Å) | | Cite: | Crystal structure of the lactonase Lmo2620 from Listeria monocytogenes
To be Published
|
|
3NQE
 
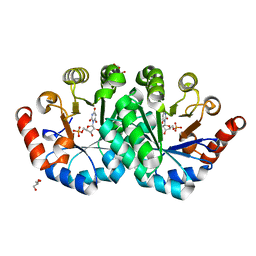 | | Crystal structure of the mutant L123N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3QY5
 
 | |
3RQF
 
 | |
3RQG
 
 | |
3Q9D
 
 | |
3QIB
 
 | | Crystal structure of the 2B4 TCR in complex with MCC/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 beta chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QEZ
 
 | | Crystal structure of the mutant T159V,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5431 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3R0D
 
 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with two zinc atoms in the active site | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Kamat, S, Hitchcock, D, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-03-07 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with two zinc atoms in the active site
To be Published
|
|
3R2P
 
 | |
3RQS
 
 | | Crystal Structure of human L-3- Hydroxyacyl-CoA dehydrogenase (EC1.1.1.35) from mitochondria at the resolution 2.0 A, Northeast Structural Genomics Consortium Target HR487, Mitochondrial Protein Partnership | | Descriptor: | GLYCEROL, Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR487
To be published
|
|
