6TN7
 
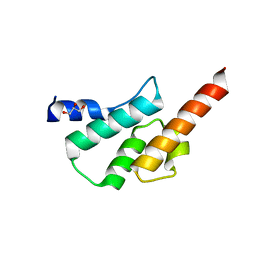 | | Crystal structure of the human Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein, GLYCEROL | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
6YI5
 
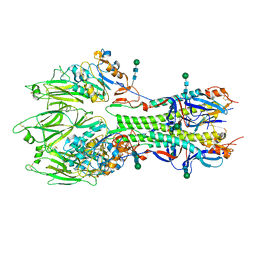 | | In-situ structure of the trimeric HEF from influenza C by flexible fitting into a cryo-ET map. | | Descriptor: | Hemagglutinin-esterase-fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Halldorsson, S, Rosenthal, P.B. | | Deposit date: | 2020-03-31 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | In situ structure and organization of the influenza C virus surface glycoprotein.
Nat Commun, 12, 2021
|
|
1KTD
 
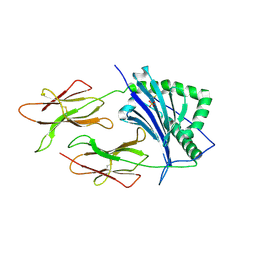 | | CRYSTAL STRUCTURE OF CLASS II MHC MOLECULE IEK BOUND TO PIGEON CYTOCHROME C PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion protein consisting of cytochrome C peptide, glycine rich linker, ... | | Authors: | Fremont, D.H, Dai, S, Chiang, H, Crawford, F, Marrack, P, Kappler, J. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of cytochrome c presentation by IE(k).
J.Exp.Med., 195, 2002
|
|
6EMH
 
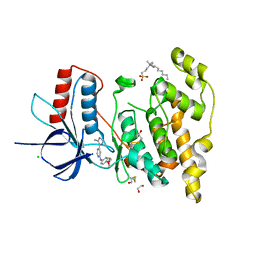 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-02 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
5XEC
 
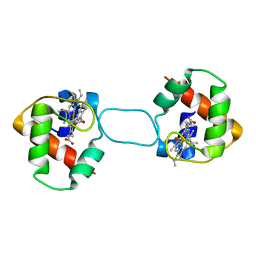 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
6LFZ
 
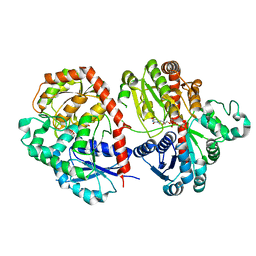 | | Crystal structure of SbCGTb in complex with UDPG | | Descriptor: | SbCGTb, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.866 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TH7
 
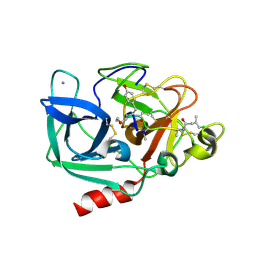 | |
2UZC
 
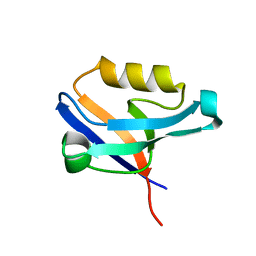 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
6TNO
 
 | | Crystal structure of the human Arc N-lobe bound to stargazin | | Descriptor: | Arc_C domain-containing protein, Chains: B,D,F | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
5QC6
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | (4~{S})-1-[1-[(2~{S})-3-[3-[3-[2-(4-methylpiperidin-1-yl)ethylsulfanyl]-4-(trifluoromethyl)phenyl]-5-methylsulfonyl-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-1-yl]-2-oxidanyl-propyl]piperidin-4-yl]-4-oxidanyl-pyrrolidin-2-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QC0
 
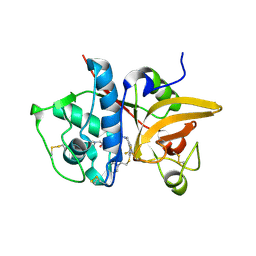 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-(dimethylamino)-1-[4-(2-oxo-2-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethyl)piperidin-1-yl]ethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCG
 
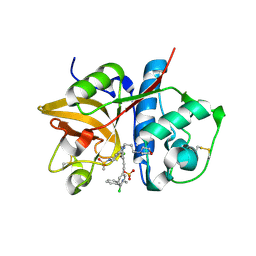 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{2-chloro-5-[2-(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethyl]phenyl}methanamine, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
4V2K
 
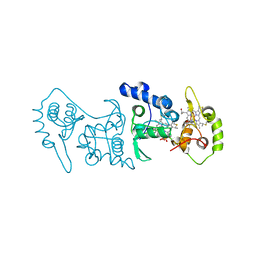 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | Descriptor: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | Authors: | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-10-10 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
5QC2
 
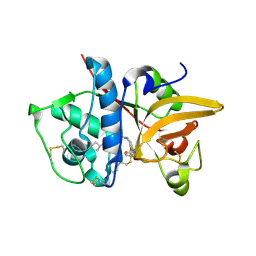 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(4-fluoropiperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-(3-hydroxypropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QBX
 
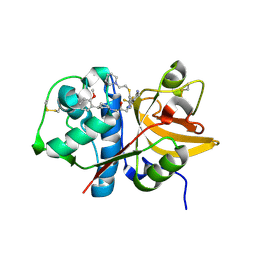 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | (2S)-1-[4-(2-methoxyphenyl)piperidin-1-yl]-3-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propan-2-ol, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCA
 
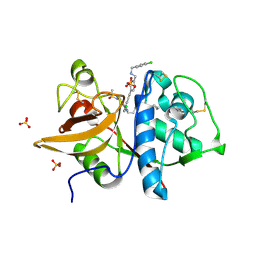 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{4-[(2-chloro-5-{1-[3-(4-cyclopropylpiperazin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}-N-[(4-chlorophenyl)methyl]methanamine, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QC7
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S, DIMETHYL SULFOXIDE, ... | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
6EXJ
 
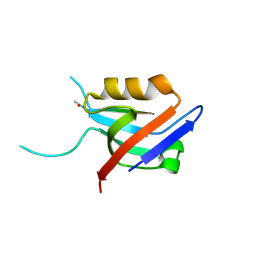 | | PDZ domain from rat Shank3 bound to the C terminus of somatostatin receptor subtype 2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 3, SSTR2 | | Authors: | Ponna, S.K, Keller, C, Ruskamo, S, Myllykoski, M, Boeckers, T.M, Kursula, P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
J. Neurochem., 145, 2018
|
|
9MT1
 
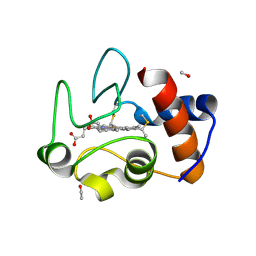 | | 1.53 A Crystal Structure of Housefly cytochrome c at pH 6.5 | | Descriptor: | Cytochrome c, ETHANOL, HEME C, ... | | Authors: | Zafreen, L, Huang, L.-S, Luemmen, P, Berry, E.A. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | Crystal Structure of Insect Cytochrome c at different pH values.
To Be Published
|
|
2IHN
 
 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
2ISO
 
 | | Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-difluoromethylene triphosphate | | Descriptor: | 2'-DEOXY-5'-O-[({[DIFLUORO(PHOSPHONO)METHYL](HYDROXY)PHOSPHORYL}OXY)(HYDROXY)PHOSPHORYL]GUANOSINE, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Sucato, C.A, Upton, T.G, Kashemirov, B.A, Martinek, V, Xiang, Y, Beard, W.A. | | Deposit date: | 2006-10-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modifying the beta,gamma Leaving-Group Bridging Oxygen Alters Nucleotide Incorporation Efficiency, Fidelity, and the Catalytic Mechanism of DNA Polymerase beta.
Biochemistry, 46, 2007
|
|
6LG0
 
 | | Crystal structure of SbCGTa in complex with UDP | | Descriptor: | SbCGTa, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2V1W
 
 | | Crystal structure of human LIM protein RIL (PDLIM4) PDZ domain bound to the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PDZ AND LIM DOMAIN PROTEIN 4, ... | | Authors: | Soundararajan, M, Shrestha, L, Pike, A.C.W, Salah, E, Burgess-Brown, N, Elkins, J, Umeano, C, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Interactions in Pdz Domain Crystal Structures Help Explain Binding Mechanisms.
Protein Sci., 19, 2010
|
|
6LOD
 
 | | Cryo-EM structure of the air-oxidized photosynthetic alternative complex III from Roseiflexus castenholzii | | Descriptor: | Cytochrome c domain-containing protein, FE3-S4 CLUSTER, Fe-S-cluster-containing hydrogenase components 1-like protein, ... | | Authors: | Shi, Y, Xin, Y.Y, Wang, C, Blankenship, R.E, Sun, F, Xu, X.L. | | Deposit date: | 2020-01-05 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the air-oxidized and dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii .
Sci Adv, 6, 2020
|
|
8BFT
 
 | |
