8OM2
 
 | | Small subunit of yeast mitochondrial ribosome in complex with METTL17/Rsm22. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM3
 
 | | Small subunit of yeast mitochondrial ribosome in complex with IF3/Aim23. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
1CU1
 
 | | CRYSTAL STRUCTURE OF AN ENZYME COMPLEX FROM HEPATITIS C VIRUS | | Descriptor: | PHOSPHATE ION, PROTEIN (PROTEASE/HELICASE NS3), ZINC ION | | Authors: | Yao, N, Weber, P.C. | | Deposit date: | 1999-08-20 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase.
Structure Fold.Des., 7, 1999
|
|
8PRW
 
 | | Cryo-EM structure of the yeast fatty acid synthase at 1.9 angstrom resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, COENZYME A, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
1D5W
 
 | | PHOSPHORYLATED FIXJ RECEIVER DOMAIN | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Birck, C, Mourey, L, Gouet, P, Fabry, B, Schumacher, J, Rousseau, P, Kahn, D, Samama, J.P. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes induced by phosphorylation of the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
8PV5
 
 | | Chaetomium thermophilum pre-60S State 8 - pre-5S rotation without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8I7J
 
 | |
1CWO
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH THR2, LEU5, D-HIV8, LEU10 CYCLOSPORIN | | Descriptor: | CYCLOSPORIN C, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-06-05 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Conformational Differences of an Immunosuppressant Peptolide in a Single Crystal and in a Crystal Complex with Human Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1CX4
 
 | |
1CZK
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 2000-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZU
 
 | | REFINED STRUCTURES OF OXIDIZED FLAVODOXIN FROM ANACYSTIS NIDULANS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Drennan, C.L, Pattridge, K.A, Weber, C.H, Metzger, A.L, Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1999-09-07 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
8IFC
 
 | | Arbekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8PVA
 
 | | Structure of bacterial ribosome determined by cryoEM at 100 keV | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L14, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1CYJ
 
 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1D09
 
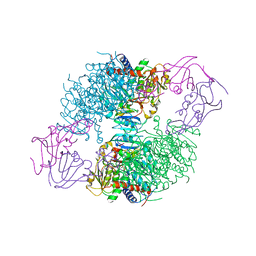 | | ASPARTATE TRANSCARBAMOYLASE COMPLEXED WITH N-PHOSPHONACETYL-L-ASPARTATE (PALA) | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Jin, L, Stec, B, Lipscomb, W.N, Kantrowitz, E.R. | | Deposit date: | 1999-09-09 | | Release date: | 2000-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanisms of catalysis and heterotropic regulation of Escherichia coli aspartate transcarbamoylase based upon a structure of the enzyme complexed with the bisubstrate analogue N-phosphonacetyl-L-aspartate at 2.1 A.
Proteins, 37, 1999
|
|
1CMK
 
 | | CRYSTAL STRUCTURES OF THE MYRISTYLATED CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE REVEAL OPEN AND CLOSED CONFORMATIONS | | Descriptor: | IODIDE ION, MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT, ... | | Authors: | Zheng, J, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Sowadski, J.M, Ten Eyck, L.F. | | Deposit date: | 1993-11-18 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations.
Protein Sci., 2, 1993
|
|
1D1Y
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1,3-PBITU (H4B FREE) | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[3-(2-CARBAMIMIDOYLSULFANYL-ETHYL)-PHENYL]-ETHYL}-ISOTHIOUREA, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
1CE5
 
 | | BOVINE PANCREAS BETA-TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ota, N, Stroupe, C, Ferreira-Da-Silva, J.M.S, Shah, S.S, Mares-Guia, M, Brunger, A.T. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Boltzmann thermodynamic integration (NBTI) for macromolecular systems: relative free energy of binding of trypsin to benzamidine and benzylamine.
Proteins, 37, 1999
|
|
1D2H
 
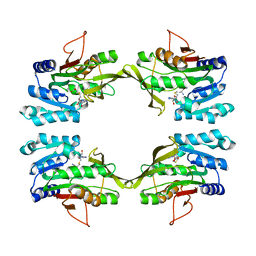 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLHOMOCYSTEINE | | Descriptor: | GLYCINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-11 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
1CEV
 
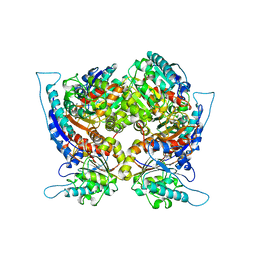 | | ARGINASE FROM BACILLUS CALDOVELOX, NATIVE STRUCTURE AT PH 5.6 | | Descriptor: | MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
8IFE
 
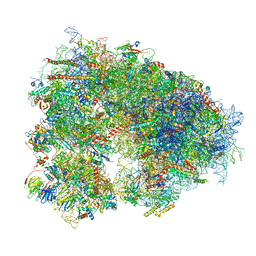 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFB
 
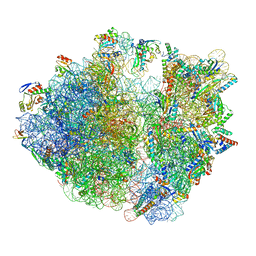 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
6VME
 
 | | Human ESCRT-I heterotetramer headpiece | | Descriptor: | Multivesicular body subunit 12A, Tumor susceptibility gene 101 protein, Vacuolar protein sorting-associated protein 28 homolog, ... | | Authors: | Flower, T.G, Hurley, J.H, Tjahjono, N. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VYS
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing a 21 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
