7BYV
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208Q with beta-1,3-galactotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Galactan 1,3-beta-galactosidase, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
2Y85
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND RCDRP | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, CHLORIDE ION, PHOSPHORIBOSYL ISOMERASE A, ... | | Authors: | Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4CON
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with citrate in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CITRIC ACID | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
7BZK
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin y1 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, IRON/SULFUR CLUSTER, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5935 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
2N4C
 
 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2FUU
 
 | |
6SXU
 
 | |
2YDF
 
 | | HUMAN SERUM ALBUMIN COMPLEXED WITH IOPHENOXIC ACID | | Descriptor: | IOPHENOXIC ACID, SERUM ALBUMIN | | Authors: | Ryan, A.J, Curry, S. | | Deposit date: | 2011-03-18 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Analysis Reveals the Structural Basis of the High-Affinity Binding of Iophenoxic Acid to Human Serum Albumin.
Bmc Struct.Biol., 11, 2011
|
|
4CSH
 
 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
4ZVK
 
 | | Reduced quinone reductase 2 in complex with ethidium | | Descriptor: | ETHIDIUM, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | Binding of DNA-Intercalating Agents to Oxidized and Reduced Quinone Reductase 2.
Biochemistry, 54, 2015
|
|
2N0Z
 
 | |
7BTS
 
 | | Structure of human beta1 adrenergic receptor bound to epinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
7BW1
 
 | | Crystal structure of Steroid 5-alpha-reductase 2 in complex with Finasteride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase 2, SULFATE ION, ... | | Authors: | Xiao, Q, Zhang, C, Wei, Z. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human steroid 5 alpha-reductase 2 with anti-androgen drug finasteride.
Res Sq, 2020
|
|
7BWP
 
 | | Crystal complex of endo-deglycosylated PcHNL5 with (R)-mandelonitrile | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
2N5Z
 
 | |
2CNR
 
 | |
2YKK
 
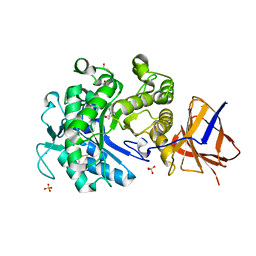 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
4ZVX
 
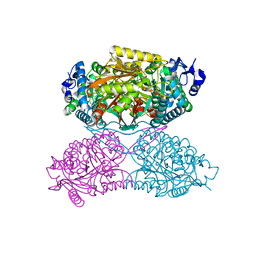 | | Structure of apo human ALDH7A1 in space group P4212 | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J. | | Deposit date: | 2015-05-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
2NC8
 
 | |
2NDI
 
 | | Solution structure of the toxin ISTX-I from Ixodes scapularis | | Descriptor: | Putative secreted salivary protein | | Authors: | Hu, K. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A sodium channel inhibitor ISTX-I with a novel structure provides a new hint at the evolutionary link between two toxin folds.
Sci Rep, 6, 2016
|
|
6SMT
 
 | | S-enantioselective imine reductase from Mycobacterium smegmatis | | Descriptor: | (2S)-2-ethylhexan-1-ol, 1,2-ETHANEDIOL, 6-phosphogluconate dehydrogenase, ... | | Authors: | Meyer, T, Zumbraegel, N, Geerds, C, Groeger, H, Niemann, H.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of an S -enantioselective Imine Reductase from Mycobacterium Smegmatis .
Biomolecules, 10, 2020
|
|
5A8E
 
 | | thermostabilised beta1-adrenoceptor with rationally designed inverse agonist 7-methylcyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-[(2S)-3-(tert-butylamino)-2-hydroxypropoxy]-7-methyl-1H-indole-2-carbonitrile, ... | | Authors: | Sato, T, Baker, J.G, Warne, T, Brown, G.A, Congreve, M, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound Beta1-Adrenergic Receptor.
Mol.Pharmacol., 88, 2015
|
|
6SUD
 
 | | Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Reducing-end xylose-releasing exo-oligoxylanase Rex8A, ... | | Authors: | Jimenez-Ortega, E, Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-09-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of the reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 deciphers its molecular specificity.
Febs J., 287, 2020
|
|
4ZX6
 
 | |
4ZY2
 
 | | X-ray crystal structure of PfA-M17 in complex with hydroxamic acid-based inhibitor 10o | | Descriptor: | CARBONATE ION, DIMETHYL SULFOXIDE, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluorobiphenyl-4-yl)ethyl]-2,2-dimethylpropanamide, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2015-05-21 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent dual inhibitors of Plasmodium falciparum M1 and M17 aminopeptidases through optimization of S1 pocket interactions.
Eur.J.Med.Chem., 110, 2016
|
|
