3KLD
 
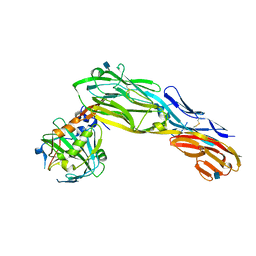 | | PTPRG CNTN4 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin 4, Receptor-type tyrosine-protein phosphatase gamma | | Authors: | Bouyain, S. | | Deposit date: | 2009-11-07 | | Release date: | 2009-12-22 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ZYI
 
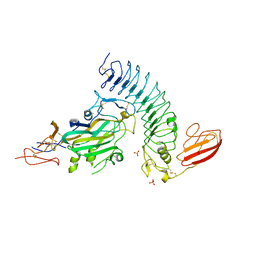 | | NetrinG2 in complex with NGL2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4, ... | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions
Embo J., 30, 2011
|
|
3ZYJ
 
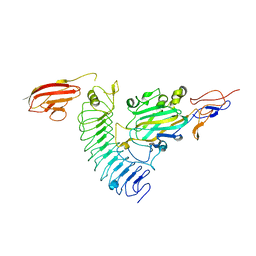 | | NetrinG1 in complex with NGL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4C, ... | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
3OJM
 
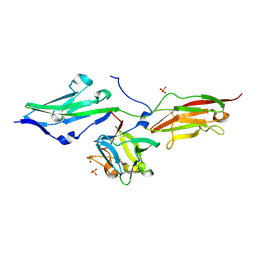 | |
3OJV
 
 | | Crystal Structure of FGF1 complexed with the ectodomain of FGFR1c exhibiting an ordered ligand specificity-determining betaC'-betaE loop | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Basic fibroblast growth factor receptor 1, Heparin-binding growth factor 1 | | Authors: | Beenken, A, Mohammadi, M. | | Deposit date: | 2010-08-23 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity in Interactions of Fibroblast Growth Factor 1 (FGF1) N Terminus with FGF Receptors Underlies Promiscuity of FGF1.
J.Biol.Chem., 287, 2012
|
|
3P40
 
 | |
3OJ2
 
 | |
3UTO
 
 | | Twitchin kinase region from C.elegans (Fn31-NL-kin-CRD-Ig26) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Castelmur, E, Barbieri, S, Mayans, O. | | Deposit date: | 2011-11-26 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of an N-terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NZI
 
 | |
3V6B
 
 | | VEGFR-2/VEGF-E complex structure | | Descriptor: | VEGF-E, Vascular endothelial growth factor receptor 2 | | Authors: | Brozzo, M.S, Leppanen, V.-M, Winkler, F.K, Kisko, K, Ballmer-Hofer, K. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization.
Blood, 119, 2012
|
|
3P3Y
 
 | |
3V2A
 
 | | VEGFR-2/VEGF-A COMPLEX STRUCTURE | | Descriptor: | Vascular endothelial growth factor A, Vascular endothelial growth factor receptor 2 | | Authors: | Brozzo, M.S, Leppanen, V.-M, Winkler, F.K, Ballmer-Hofer, K. | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization.
Blood, 119, 2012
|
|
1EVT
 
 | | CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 1), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 1), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1FQ9
 
 | | CRYSTAL STRUCTURE OF A TERNARY FGF2-FGFR1-HEPARIN COMPLEX | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-beta-L-altropyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 2, ... | | Authors: | Schlessinger, J, Plotnikov, A.N, Ibrahimi, O.A, Eliseenkova, A.V, Yeh, B.K, Yayon, A, Linhardt, R.J, Mohammadi, M. | | Deposit date: | 2000-09-04 | | Release date: | 2000-09-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization.
Mol.Cell, 6, 2000
|
|
1RY7
 
 | | Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 | | Descriptor: | Fibroblast growth factor receptor 3, Heparin-binding growth factor 1 | | Authors: | Olsen, S.K, Ibrahimi, O.A, Raucci, A, Zhang, F, Eliseenkova, A.V, Yayon, A, Basilico, C, Linhardt, R.J, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1VCA
 
 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
1HCF
 
 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | Authors: | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7YSW
 
 | | Cryo-EM Structure of FGF23-FGFR4-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
6FF3
 
 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L1 with Human IGF-I | | Descriptor: | Insulin-like growth factor I, Neural/ectodermal development factor IMP-L2 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D, Viola, C.M. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
6FEY
 
 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L2 with Drosophila DILP5 insulin | | Descriptor: | Neural/ectodermal development factor IMP-L2, Probable insulin-like peptide 5 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
6G2T
 
 | | human cardiac myosin binding protein C C1 Ig-domain bound to native cardiac thin filament | | Descriptor: | Actin, cytoplasmic 2, Myosin-binding protein C, ... | | Authors: | Risi, C, Belknap, B, Forgacs, E, Harris, S.P, Schroder, G.F, White, H.D, Galkin, V.E. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament.
Structure, 26, 2018
|
|
