2Y4L
 
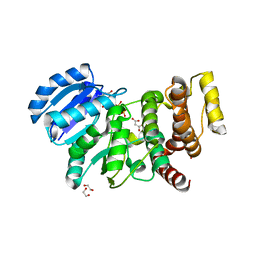 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH Manganese and GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MALONATE ION, ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
6C1M
 
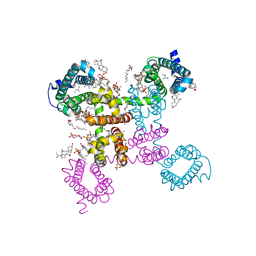 | | NavAb NormoPP mutant | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-METHYLGUANIDINE, CHAPSO, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
4ZAX
 
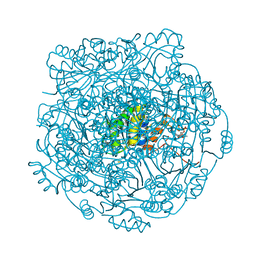 | | Structure of UbiX in complex with oxidised prenylated FMN (radical) | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
6BI1
 
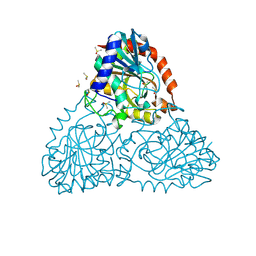 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with (2R)-2-amino-3-(benzyloxy)propan-1-ol | | Descriptor: | DIMETHYL SULFOXIDE, O-benzyl-L-serine, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with (2R)-2-amino-3-(benzyloxy)propan-1-ol
To Be Published
|
|
7BIV
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1,2-ETHANEDIOL, 6-[[(1~{R})-1-(4-chlorophenyl)-7-fluoranyl-1-[[1-(hydroxymethyl)cyclopropyl]methoxy]-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-2-yl]methyl]pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BIR
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1-[[(1~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-1-(4-chlorophenyl)-7-fluoranyl-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-1-yl]oxymethyl]cyclopropane-1-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
5ZJ3
 
 | |
7FYJ
 
 | | Crystal Structure of human FABP4 in complex with 5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one, i.e. SMILES c1(cccc(c1C)Cl)OCC1=CC(=O)N2N=C(c3ccccc3)N=C2N1 with IC50=0.061 microM | | Descriptor: | (8S)-5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4B90
 
 | | Crystal structure of WT human CRMP-5 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
7FZ7
 
 | | Crystal Structure of human FABP4 in complex with 2-[2-[(2-chlorophenyl)methyl]-1-hydroxycyclohexyl]acetic acid, i.e. SMILES [C@]1([C@@H](Cc2c(cccc2)Cl)CCCC1)(CC(=O)O)O with IC50=1.7 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, SULFATE ION, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Dostert, P, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8K53
 
 | |
7G1S
 
 | | Crystal Structure of human FABP4 in complex with 3-(3,4-dichlorophenyl)-3,7a-dihydro-1H-imidazo[1,5-c][1,3]thiazole-5,7-dione | | Descriptor: | (3R,4R,7aS)-3-(3,4-dichlorophenyl)-1H,3H-imidazo[1,5-c][1,3]thiazole-5,7(6H,7aH)-dione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Zeller, P, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1NMV
 
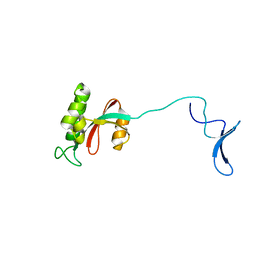 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
6LR6
 
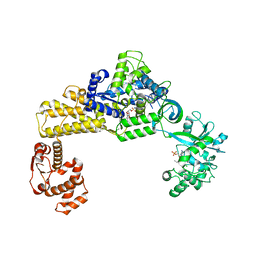 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6BUT
 
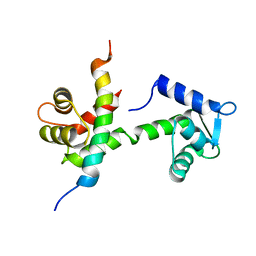 | |
7FXK
 
 | | Crystal Structure of human FABP4 in complex with 5-benzyl-4-(2,5-dimethylpyrrol-1-yl)-1,2,4-triazole-3-thiol, i.e. SMILES N1(N2C(=CC=C2C)C)C(=NN=C1S)Cc1ccccc1 with IC50=0.462 microM | | Descriptor: | 5-benzyl-4-(2,5-dimethyl-1H-pyrrol-1-yl)-4H-1,2,4-triazole-3-thiol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
2B8R
 
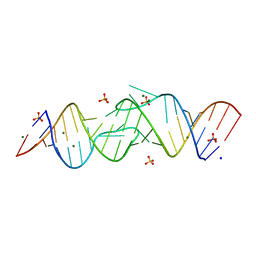 | | Structure oF HIV-1(LAI) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
Nat.Struct.Biol., 8, 2001
|
|
6LSM
 
 | | Tubulin Polymerization Inhibitors | | Descriptor: | 2-(4-methylphenyl)-7-(3,4,5-trimethoxyphenyl)pyrazolo[1,5-a]pyrimidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gang, L, Wang, Y.X, Chen, J.J. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Design, Synthesis, and Bioevaluation of Pyrazolo[1,5-a]Pyrimidine Derivatives as Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site with Potent Anticancer Activities
To Be Published
|
|
7FYK
 
 | | Crystal Structure of human FABP4 in complex with 5-phenylmethoxy-4-prop-2-enyl-1,2,4-triazole-3-thiol, i.e. SMILES N1(C(=NN=C1S)OCc1ccccc1)CC=C with IC50=1.3 microM | | Descriptor: | 5-(benzyloxy)-4-(prop-2-en-1-yl)-4H-1,2,4-triazole-3-thiol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5ZJ6
 
 | | Crystal structure of HCK kinase complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2018-03-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Phosphorylated and non-phosphorylated HCK kinase domains produced by cell-free protein expression.
Protein Expr. Purif., 150, 2018
|
|
6TEL
 
 | | Crystal structure of Dot1L in complex with an inhibitor (compound 10). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, POTASSIUM ION, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
7FYP
 
 | | Crystal Structure of human FABP4 in complex with (1S,2S)-2-[(1R,2S,5R)-5-methyl-2-propan-2-ylcyclohexyl]oxycarbonylcyclopropane-1-carboxylic acid, i.e. SMILES [C@H]1([C@H](C1)C(=O)O)C(=O)O[C@H]1[C@@H](CC[C@H](C1)C)C(C)C with IC50=2.6 microM | | Descriptor: | (1S,2S)-2-({[(1R,2S,5R)-5-methyl-2-(propan-2-yl)cyclohexyl]oxy}carbonyl)cyclopropane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Woltering, T, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5TIJ
 
 | | Structure of Human Enolase 2 with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonate (purified enantiomer) | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Eradication of ENO1-deleted Glioblastoma through Collateral Lethality
Biorxiv, 2019
|
|
4Z3M
 
 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-03-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
7YR5
 
 | |
