1T6C
 
 | | Structural characterization of the Ppx/GppA protein family: crystal structure of the Aquifex aeolicus family member | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kristensen, O, Laurberg, M, Liljas, A, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family
Biochemistry, 43, 2004
|
|
1T3Z
 
 | | Formyl-CoA Tranferase mutant Asp169 to Ser | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1TJC
 
 | | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | | Descriptor: | Prolyl 4-hydroxylase alpha-1 subunit | | Authors: | Pekkala, M, Hieta, R, Bergmann, U, Kivirikko, K.I, Wierenga, R.K, Myllyharju, J. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Peptide-Substrate-binding Domain of Collagen Prolyl 4-Hydroxylases Is a Tetratricopeptide Repeat Domain with Functional Aromatic Residues.
J.Biol.Chem., 279, 2004
|
|
1TEF
 
 | | Crystal structure of the spinach plastocyanin mutants G8D/K30C/T69C and K30C/T69C- a study of the effect on crystal packing and thermostability from the introduction of a novel disulfide bond | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Plastocyanin, ... | | Authors: | Okvist, M, Jacobson, F, Jansson, H, Hansson, O, Sjolin, L. | | Deposit date: | 2004-05-25 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 and 1.96 resolution, respectively.
To be Published
|
|
1TJ7
 
 | | Structure determination and refinement at 2.44 A resolution of Argininosuccinate lyase from E. coli | | Descriptor: | Argininosuccinate lyase, GLYCEROL, PHOSPHATE ION | | Authors: | Bhaumik, P, Koski, M.K, Bergman, U, Wierenga, R.K. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure determination and refinement at 2.44 A resolution of argininosuccinate lyase from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8RDR
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8g | | Descriptor: | (5~{S})-3-(2-nitrophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDS
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8i | | Descriptor: | (5~{S})-3-(2-methoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDT
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8j | | Descriptor: | (5~{S})-3-(2,3,4-trimethoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDQ
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8b | | Descriptor: | (5S)-3-(2,4-dichlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8RDP
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8a | | Descriptor: | (5~{S})-3-(2-chlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
1UHL
 
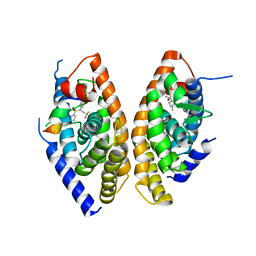 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
7NKG
 
 | |
7NWL
 
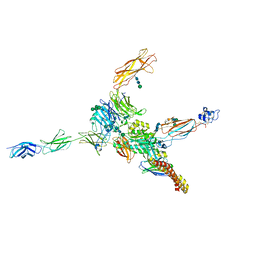 | | Cryo-EM structure of human integrin alpha5beta1 (open form) in complex with fibronectin and TS2/16 Fv-clasp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-5, ... | | Authors: | Schumacher, S, Dedden, D, Vazquez Nunez, R, Matoba, K, Takagi, J, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into integrin alpha 5 beta 1 opening by fibronectin ligand.
Sci Adv, 7, 2021
|
|
1USB
 
 | |
1W6I
 
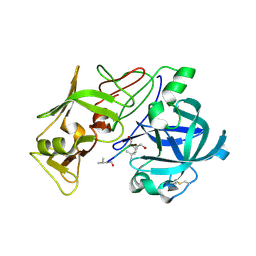 | | plasmepsin II-pepstatin A complex | | Descriptor: | PEPSTATIN, PLASMEPSIN 2 PRECURSOR | | Authors: | Lindberg, J, Johansson, P.-O, Rosenquist, A, Kvarnstroem, I, Vrang, L, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-18 | | Release date: | 2006-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Study of a Novel Inhibitor with Bulky P1 Side Chain in Complex with Plasmepsin II -Implications for Drug Design
To be Published
|
|
1WBM
 
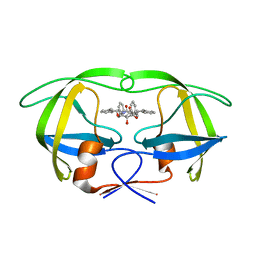 | | HIV-1 protease in complex with symmetric inhibitor, BEA450 | | Descriptor: | (2R,3R,4R,5R)-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]-2,5-BIS(2-PHENYLETHYL)HEXANEDIAMIDE, POL PROTEIN (FRAGMENT) | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease in Complex with Symmetric Inhibitor, Bea450
To be Published
|
|
1W4Y
 
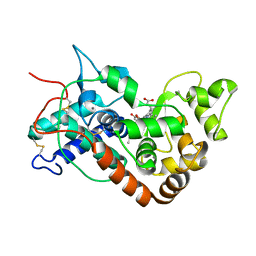 | | Ferrous horseradish peroxidase C1A in complex with carbon monoxide | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
1WB7
 
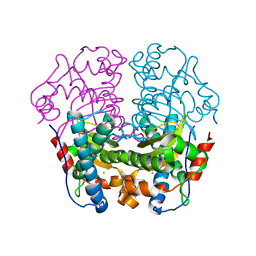 | | Iron Superoxide Dismutase (Fe-SOD) From The Hyperthermophile Sulfolobus Solfataricus. Crystal Structure of the Y41F mutant. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE [FE] | | Authors: | Gogliettino, M.A, Tanfani, F, Scire, A, Ursby, T, Adinolfi, B.S, Cacciamani, T, De Vendittis, E. | | Deposit date: | 2004-10-31 | | Release date: | 2004-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Role of Tyr41 and His155 in the Functional Properties of Superoxide Dismutase from the Archaeon Sulfolobus Solfataricus
Biochemistry, 43, 2004
|
|
1W4W
 
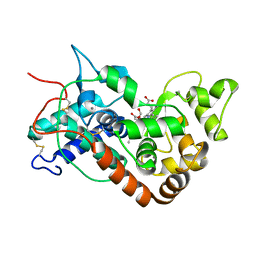 | | Ferric horseradish peroxidase C1A in complex with formate | | Descriptor: | CALCIUM ION, FORMIC ACID, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
1WBK
 
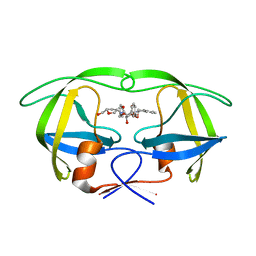 | |
1USI
 
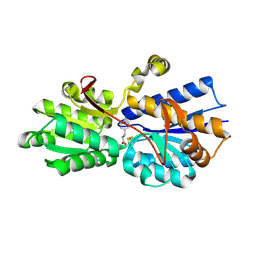 | | L-leucine-binding protein with phenylalanine bound | | Descriptor: | LEUCINE-SPECIFIC BINDING PROTEIN, PHENYLALANINE | | Authors: | Magnusson, U, Salopek-Sondi, B, Luck, L.A, Mowbray, S.L. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Leucine-Binding Protein Illustrate Conformational Changes and the Basis of Ligand Specificity
J.Biol.Chem., 279, 2004
|
|
1XS4
 
 | | dCTP deaminase from Escherichia coli- E138A mutant enzyme in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
