6TP5
 
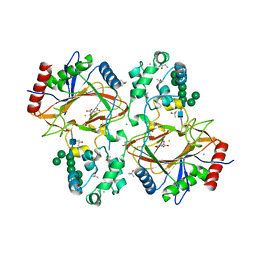 | | Crystal structure of human Transmembrane prolyl 4-hydroxylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Myllykoski, M, Sutinen, A, Koski, M.K, Kallio, J.P, Raasakka, A, Myllyharju, J, Wierenga, R.K, Koivunen, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of transmembrane prolyl 4-hydroxylase reveals unique organization of EF and dioxygenase domains.
J.Biol.Chem., 296, 2020
|
|
2ALB
 
 | |
1TJC
 
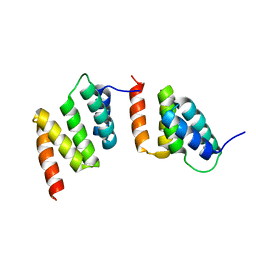 | | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | | Descriptor: | Prolyl 4-hydroxylase alpha-1 subunit | | Authors: | Pekkala, M, Hieta, R, Bergmann, U, Kivirikko, K.I, Wierenga, R.K, Myllyharju, J. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Peptide-Substrate-binding Domain of Collagen Prolyl 4-Hydroxylases Is a Tetratricopeptide Repeat Domain with Functional Aromatic Residues.
J.Biol.Chem., 279, 2004
|
|
2V4A
 
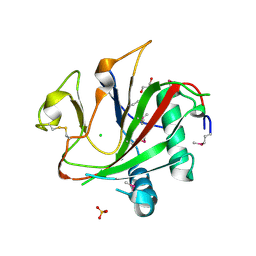 | | Crystal structure of the SeMet-labeled prolyl-4 hydroxylase (P4H) type I from green algae Chlamydomonas reinhardtii. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
2V5F
 
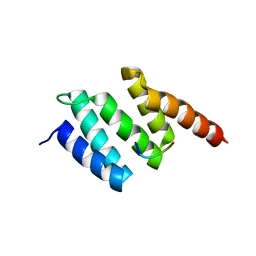 | | Crystal structure of wild type peptide-binding domain of human type I collagen prolyl 4-hydroxylase. | | Descriptor: | HEXA-HISTIDINE PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Pekkala, M, Hieta, R, Kivirikko, K, Myllyharju, J, Wierenga, R. | | Deposit date: | 2008-10-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Wild Type Peptide-Binding Domain of Human Type I Collagen Prolyl 4- Hydroxylase.
To be Published
|
|
2JIJ
 
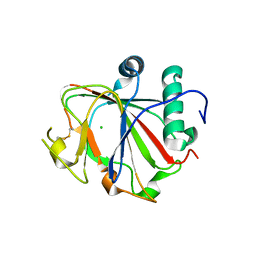 | | Crystal structure of the apo form of Chlamydomonas reinhardtii prolyl- 4 hydroxylase type I | | Descriptor: | CHLORIDE ION, PROLYL-4 HYDROXYLASE | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
2JIG
 
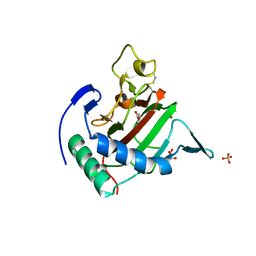 | | Crystal structure of Chlamydomonas reinhardtii prolyl-4 hydroxylase type I complexed with zinc and pyridine-2,4-dicarboxylate | | Descriptor: | GLYCEROL, PROLYL-4 HYDROXYLASE, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
