1ML1
 
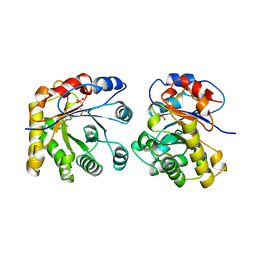 | | PROTEIN ENGINEERING WITH MONOMERIC TRIOSEPHOSPHATE ISOMERASE: THE MODELLING AND STRUCTURE VERIFICATION OF A SEVEN RESIDUE LOOP | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Thanki, N, Zeelen, J.P, Mathieu, M, Jaenicke, R, Abagyan, R.A, Wierenga, R, Schliebs, W. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein engineering with monomeric triosephosphate isomerase (monoTIM): the modelling and structure verification of a seven-residue loop.
Protein Eng., 10, 1997
|
|
1AMK
 
 | | LEISHMANIA MEXICANA TRIOSE PHOSPHATE ISOMERASE | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSE PHOSPHATE ISOMERASE | | Authors: | Williams, J.C, Wierenga, R. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power.
Protein Eng., 12, 1999
|
|
2PTK
 
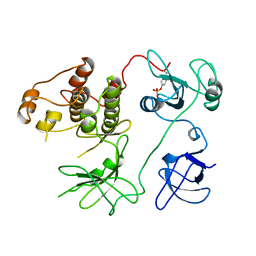 | | CHICKEN SRC TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Williams, J.C, Wierenga, R. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions
J.Mol.Biol., 274, 1997
|
|
7R3D
 
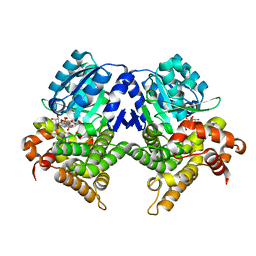 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND GLYCEROL (Absence of Nicotinamide ring) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, Lactaldehyde reductase | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-07 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
5BYV
 
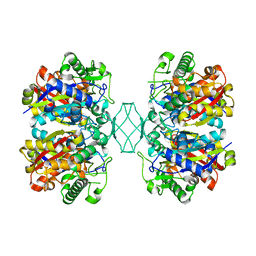 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5F0V
 
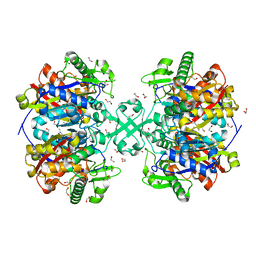 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-11-28 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5F38
 
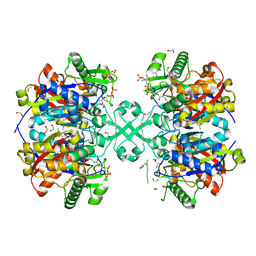 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase, COENZYME A, ... | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-12-02 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4B3H
 
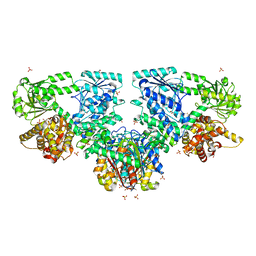 | |
4ZRC
 
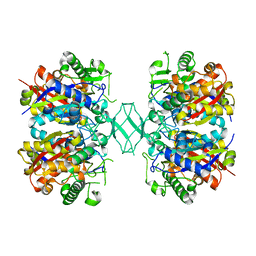 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | Descriptor: | Beta-ketothiolase | | Authors: | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7R5T
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-11 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
2V5F
 
 | | Crystal structure of wild type peptide-binding domain of human type I collagen prolyl 4-hydroxylase. | | Descriptor: | HEXA-HISTIDINE PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Pekkala, M, Hieta, R, Kivirikko, K, Myllyharju, J, Wierenga, R. | | Deposit date: | 2008-10-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Wild Type Peptide-Binding Domain of Human Type I Collagen Prolyl 4- Hydroxylase.
To be Published
|
|
2X16
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2009-12-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1U
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1S
 
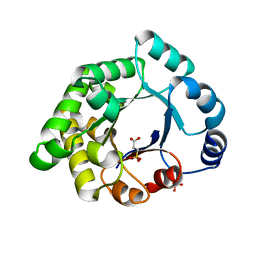 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1R
 
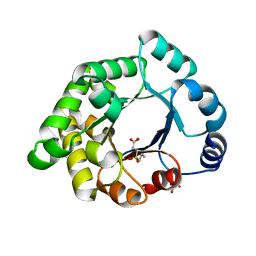 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-(PROPYLSULFONYL)PROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1T
 
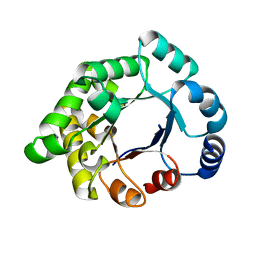 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X2G
 
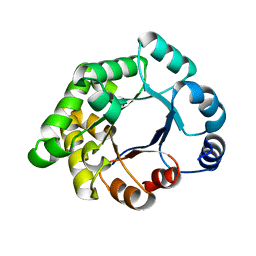 | | CRYSTALLOGRAPHIC BINDING STUDIES WITH AN ENGINEERED MONOMERIC VARIANT OF TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1SHF
 
 | |
