1P5Y
 
 | |
1P5W
 
 | |
1OIV
 
 | | X-ray structure of the small G protein Rab11a in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, RAS-RELATED PROTEIN RAB-11A, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
1Q35
 
 | | Crystal Structure of Pasteurella haemolytica Apo Ferric ion-Binding Protein A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, iron binding protein FbpA | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Snell, G, Scheibe, D, Williams, P.A, Kirby, S, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins
J.Biol.Chem., 278, 2003
|
|
5W1M
 
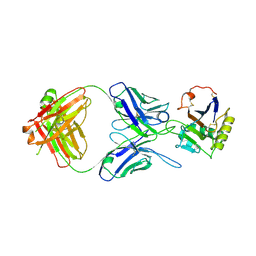 | | MACV GP1 CR1-07 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR1-07 Fab heavy chain, ... | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
2CAM
 
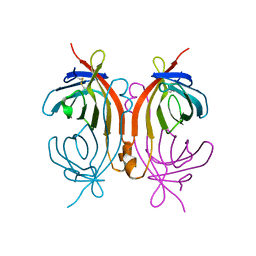 | | AVIDIN MUTANT (K3E,K9E,R26D,R124L) | | Descriptor: | AVIDIN | | Authors: | Rosano, C, Arosio, P, Bolognesi, M. | | Deposit date: | 1998-03-27 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical characterization and crystal structure of a recombinant hen avidin and its acidic mutant expressed in Escherichia coli.
Eur.J.Biochem., 256, 1998
|
|
5W1G
 
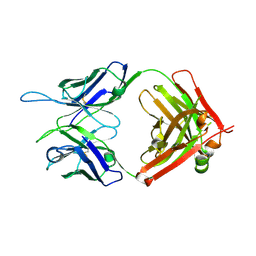 | | CR1-07 unliganded Fab | | Descriptor: | CR1-07 Fab heavy chain, CR1-07 Fab light chain | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
6E7P
 
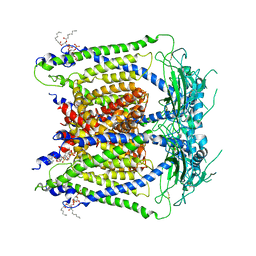 | | cryo-EM structure of human TRPML1 with PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
5WJ9
 
 | | Human TRPML1 channel structure in agonist-bound open conformation | | Descriptor: | 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Human TRPML1 channel structures in open and closed conformations.
Nature, 550, 2017
|
|
6E7Y
 
 | | cryo-EM structure of human TRPML1 with PI45P2 | | Descriptor: | Mucolipin-1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
5TCX
 
 | | Crystal structure of human tetraspanin CD81 | | Descriptor: | CD81 antigen, CHOLESTEROL | | Authors: | Zimmerman, B, McMillan, B.J, Seegar, T.C.M, Kruse, A.C, Blacklow, S.C. | | Deposit date: | 2016-09-16 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket.
Cell, 167, 2016
|
|
6E7Z
 
 | | cryo-EM structure of human TRPML1 with ML-SA1 and PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
5WJ5
 
 | |
5W1K
 
 | | JUNV GP1 CR1-10 Fab CR1-28 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR1-10 Fab heavy chain, ... | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
2F9L
 
 | | 3D structure of inactive human Rab11b GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAB11B, ... | | Authors: | Scapin, S.M.N, Guimaraes, B.G, Zanchin, N.I.T. | | Deposit date: | 2005-12-06 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform.
J.Struct.Biol., 154, 2006
|
|
2F9M
 
 | | 3D structure of active human Rab11b GTPase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Scapin, S.M.N, Guimaraes, B.G, Zanchin, N.I.T. | | Deposit date: | 2005-12-06 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform.
J.Struct.Biol., 154, 2006
|
|
7KQ8
 
 | | Structure of iron bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-11-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
7MGL
 
 | | Structure of human TRPML1 with ML-SI3 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Mucolipin-1, N-{(1S,2S)-2-[4-(2-methoxyphenyl)piperazin-1-yl]cyclohexyl}benzenesulfonamide | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic insights into ML-SI3 mediated human TRPML1 inhibition.
Structure, 29, 2021
|
|
7M8H
 
 | | Structure of Memo1 C244S metal binding site mutant at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2021-03-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
3FF3
 
 | |
3FEC
 
 | | Crystal structure of human Glutamate Carboxypeptidase III (GCPIII/NAALADase II), pseudo-unliganded | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Barinka, C, Lubkowski, J, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
7L5C
 
 | | Structure of copper bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, GLYCEROL, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
3FEE
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FED
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with a transition state analog of Glu-Glu | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-amino-3-carboxypropyl](hydroxy)phosphoryl]methyl}pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
6IRL
 
 | |
