6E7J
 
 | | HIV-1 wild type protease with GRL-042-17A, 3-phenylhexahydro-2h-cyclopenta[d]oxazol-2-one with a bicyclic oxazolidinone scaffold as the P2 ligand | | Descriptor: | (3aS,5R,6aR)-2-oxo-3-phenylhexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
1S56
 
 | | Crystal Structure of "Truncated" Hemoglobin N (HbN) from Mycobacterium tuberculosis, Soaked with Xe Atoms | | Descriptor: | CYANIDE ION, HEME C, Hemoglobin-like protein HbN, ... | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-20 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Heme-ligand tunneling in group I truncated hemoglobins
J.Biol.Chem., 279, 2004
|
|
6E9A
 
 | | HIV-1 WILD TYPE PROTEASE WITH GRL-034-17A, (3aS, 5R, 6aR)-2-OXOHEXAHYD CYCLOPENTA[D]-5-OXAZOLYL URETHANE WITH A BICYCLIC OXAZOLIDINONE SCAFF AS THE P2 LIGAND | | Descriptor: | (3aS,5R,6aR)-2-oxohexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
5U8C
 
 | |
2HEO
 
 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | Authors: | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
2D3T
 
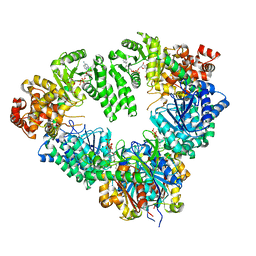 | | Fatty Acid beta-oxidation multienzyme complex from Pseudomonas Fragi, Form V | | Descriptor: | 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, Fatty oxidation complex alpha subunit, ... | | Authors: | Tsuchiya, D, Shimizu, N, Ishikawa, M, Suzuki, Y, Morikawa, K. | | Deposit date: | 2005-10-01 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-Induced Domain Rearrangement of Fatty Acid beta-Oxidation Multienzyme Complex
Structure, 14, 2006
|
|
5N65
 
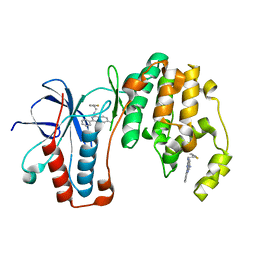 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N64
 
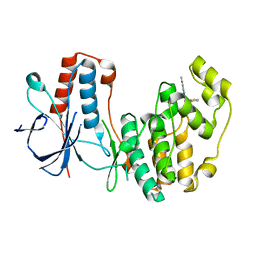 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9g | | Descriptor: | 2-phenyl-~{N}4-(thiophen-2-ylmethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
7XYZ
 
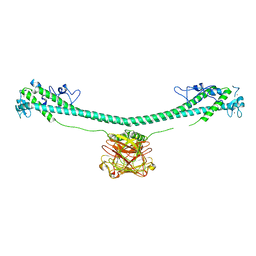 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.62 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XZ2
 
 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7A9Z
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 1 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AA1
 
 | | Structural comparison of cellular retinoic acid binding proteins I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7A9Y
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | Cellular retinoic acid-binding protein 1, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2DM5
 
 | | Thermodynamic Penalty Arising From Burial of a Ligand Polar Group Within a Hydrophobic Pocket of a Protein Receptor | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTANE-1,8-DIOL | | Authors: | Barratt, E, Bronowska, A, Vondrasek, J, Bingham, R, Phillips, S, Homans, S.W. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor
J.Mol.Biol., 362, 2006
|
|
7AA0
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | (~{E})-3-[4-(4,4-dimethyl-1-propan-2-yl-2,3-dihydroquinolin-6-yl)phenyl]prop-2-enoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3T5C
 
 | |
5E4K
 
 | | Structure of ligand binding region of uPARAP at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
3T5A
 
 | |
5HA1
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with retinylamine | | Descriptor: | (2~{E},4~{E},6~{E},8~{E})-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraen-1-amine, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
5N68
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9m | | Descriptor: | 2-(4-morpholin-4-ylphenyl)-~{N}4-(2-phenylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5H9A
 
 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
5N67
 
 | |
7V8G
 
 | | Crystal structure of HOIP RING1 domain bound to IpaH1.4 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, RING-type E3 ubiquitin transferase, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5EW6
 
 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
7XYY
 
 | | TRIM E3 ubiquitin ligase WT | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
