3L3U
 
 | | Crystal structure of the HIV-1 integrase core domain to 1.4A | | Descriptor: | POL polyprotein, SULFATE ION | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.
Febs Lett., 584, 2010
|
|
5C9M
 
 | | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.362 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution.
To Be Published
|
|
4DCQ
 
 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
5CEP
 
 | |
3KVV
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
1PLW
 
 | |
3KY8
 
 | |
6A1C
 
 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FBM
 
 | | Crystal Structure of Histone Like Protein (HLP) from Streptococcus mutans Refined to 1.9 A Resolution | | Descriptor: | DNA-binding protein HU | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, O'Neil, P, Biswas, I. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of histone-like protein from Streptococcus mutans refined to 1.9 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
1XY4
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XXE
 
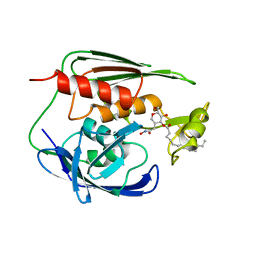 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
4LTA
 
 | |
1RPB
 
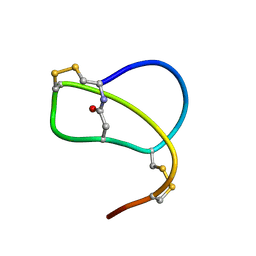 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1MKI
 
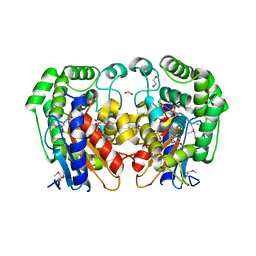 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
5G09
 
 | | The crystal structure of a S-selective transaminase from Bacillus megaterium bound with R-alpha-methylbenzylamine | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
4M5O
 
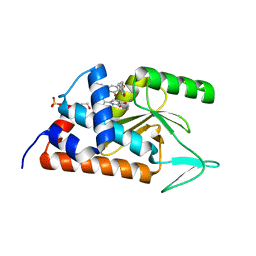 | | 3-HYDROXY-6-PHENYL-1,2-DIHYDROPYRIDIN-2-ONE bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-phenylpyridin-2(5H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
6NFE
 
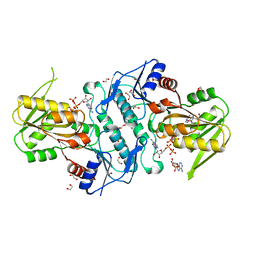 | |
4LWG
 
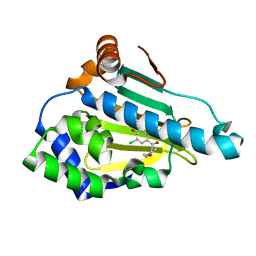 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
6A6L
 
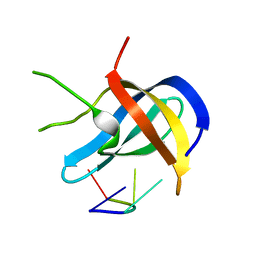 | |
4PNL
 
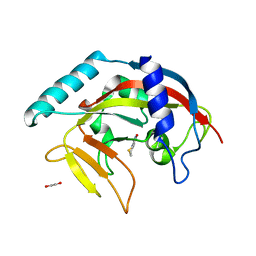 | | Crystal structure of TNKS-2 in complex with DR2313. | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1XY9
 
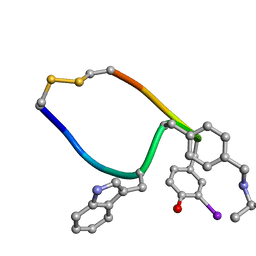 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
4EZ2
 
 | |
1XY5
 
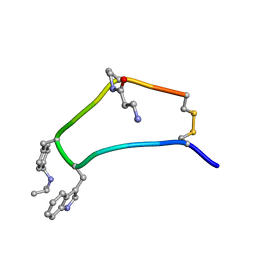 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
2ZDU
 
 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(4-{[6-bromo-3-(methoxycarbonyl)-1-oxo-4-phenylisoquinolin-2(1H)-yl]methyl}phenyl)amino]-4-oxobutanoic acid, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Ohra, T, Itoh, F, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (1)
Bioorg.Med.Chem., 16, 2008
|
|
