8G36
 
 | | Crystal structure of F182L-CYP199A4 in complex with terephthalic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
8G35
 
 | | Crystal structure of F182L-CYP199A4 in complex with (S)-4-(2-hydroxy-3-oxobutan-2-yl)benzoic acid | | Descriptor: | 4-[(2S)-2-hydroxy-3-oxobutan-2-yl]benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bell, S.G, Bruning, J.B. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
8G30
 
 | |
8G2Y
 
 | | Cryo-EM structure of ADGRF1 coupled to miniGs/q | | Descriptor: | Adhesion G-protein-coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jones, D, Rawson, S, Blacklow, S. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Tethered agonist activated ADGRF1 structure and signalling analysis reveal basis for G protein coupling.
Nat Commun, 14, 2023
|
|
8G2Z
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain CU428 | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
8CGG
 
 | |
8CGH
 
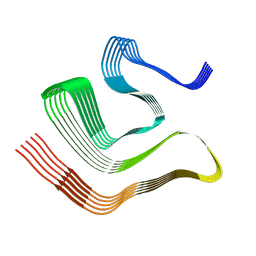 | |
8CG3
 
 | |
8G28
 
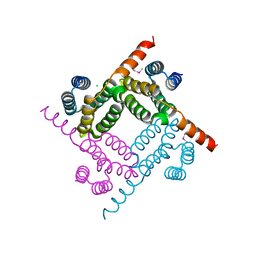 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | Descriptor: | ATPase, AAA family, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
8G24
 
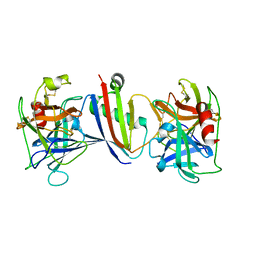 | |
8G25
 
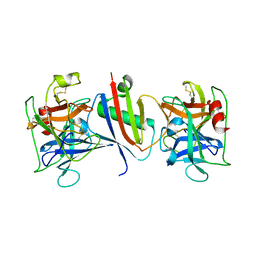 | |
8G26
 
 | |
8G1W
 
 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor VD4162B | | Descriptor: | CHLORIDE ION, Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(KCM), Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(THZ), ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism-Based Macrocyclic Inhibitors of Serine Proteases.
J.Med.Chem., 67, 2024
|
|
8G1V
 
 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor MM1132-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, N~2~-{[3-(acetamidomethyl)phenyl]acetyl}-N-[(2S)-1-(1,3-benzothiazol-2-yl)-5-carbamimidamido-1,1-dihydroxypentan-2-yl]-L-leucinamide, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Use of protease substrate specificity screening in the rational design of selective protease inhibitors with unnatural amino acids: Application to HGFA, matriptase, and hepsin.
Protein Sci., 33, 2024
|
|
8I7R
 
 | | In situ structure of axonemal doublet microtubules in mouse sperm with 48-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
8I7W
 
 | | Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8I7V
 
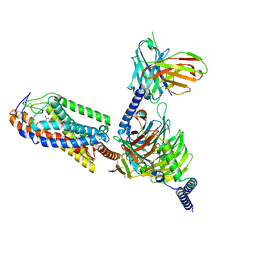 | | Cryo-EM structure of Acipimox bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8CF4
 
 | |
8CE9
 
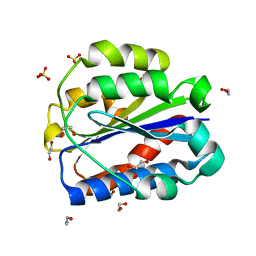 | | Crystal structure of human Cd11b I domain in C121 space group | | Descriptor: | 1,2-ETHANEDIOL, Integrin alpha-M, SULFATE ION | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human Cd11b I domain in C121 space group
To Be Published
|
|
8I7O
 
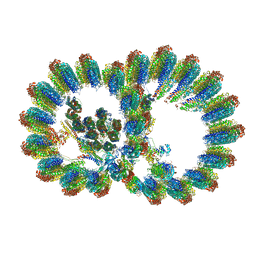 | | In situ structure of axonemal doublet microtubules in mouse sperm with 16-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 276, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
8CEA
 
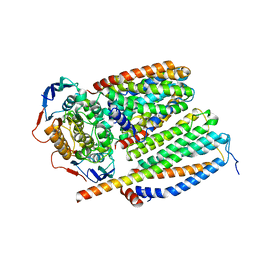 | | Cytochrome c maturation complex CcmABCD, E154Q | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE1
 
 | | Cytochrome c maturation complex CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CDW
 
 | |
8CE8
 
 | | Cytochrome c maturation complex CcmABCDE | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Cytochrome c-type biogenesis protein CcmE, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
8CE5
 
 | | Cytochrome c maturation complex CcmABCD, E154Q, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
