7S4G
 
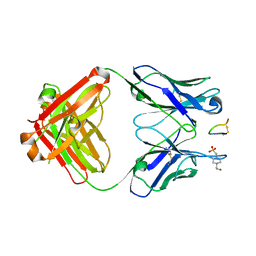 | | Fab fragment bound to the Cter peptide of Ly6G6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lymphocyte antigen 6 complex locus protein G6d, ... | | Authors: | Rouge, L, Lupardus, P. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Anti-LY6G6D/CD3 T-Cell-Dependent Bispecific Antibody for the Treatment of Colorectal Cancer.
Mol.Cancer Ther., 21, 2022
|
|
5EAJ
 
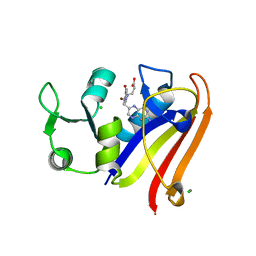 | | Crystal structure of DHFR in 0% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
4P3Q
 
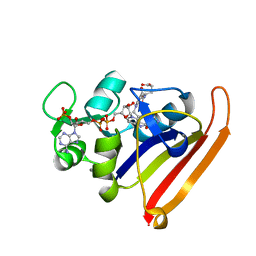 | | Room-temperature WT DHFR, time-averaged ensemble | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Keedy, D.A, van den Bedem, H, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
8GYK
 
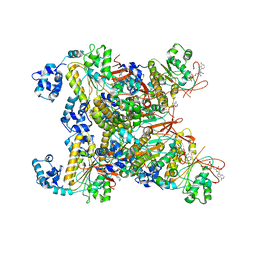 | | CryoEM structure of the RAD51_ADP filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 1, MAGNESIUM ION | | Authors: | Miki, Y, Luo, S.C, Ho, M.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | A RAD51-ADP double filament structure unveils the mechanism of filament dynamics in homologous recombination.
Nat Commun, 14, 2023
|
|
3ZFI
 
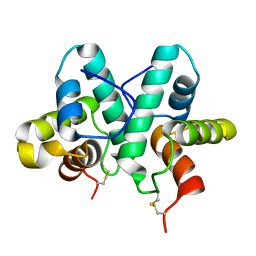 | | Rap1a protein (SMA2260) from Serratia marcescens | | Descriptor: | RAP1A PROTEIN | | Authors: | Srikannathasan, V, O'Rourke, P.E.F, Rao, V.A, English, G, Coulthurst, S.J, Hunter, W.N. | | Deposit date: | 2012-12-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis for Type Vi Secreted Peptidoglycan Dl-Endopeptidase Function, Specificity and Neutralization in Serratia Marcescens
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZEV
 
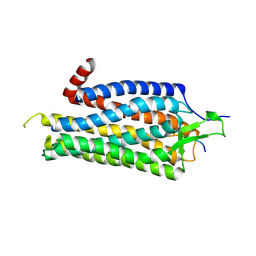 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5CC9
 
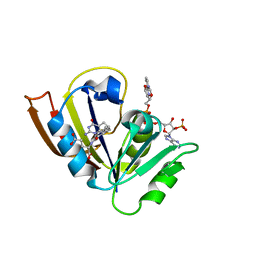 | |
5DNR
 
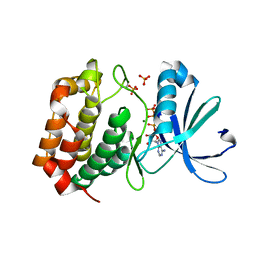 | | Aurora A Kinase in complex with ATP in space group P41212 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G, Huggins, D, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A. | | Deposit date: | 2015-09-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5E9Z
 
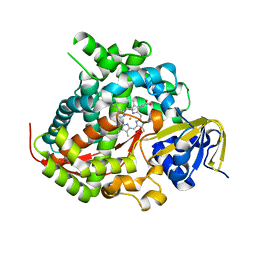 | | Cytochrome P450 BM3 mutant M11 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, FE (II) ION, ... | | Authors: | Capoferri, L, Leth, R, ter Haar, E, Mohanty, A.K, Grootenhuis, D.J, Vottero, E, Commandeur, J.N.M, Vermeulen, N.P.E, Jorgensen, F.S, Olsen, L, Geerke, D.P. | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into regioselective metabolism of mefenamic acid by cytochrome P450 BM3 mutants through crystallography, docking, molecular dynamics, and free energy calculations.
Proteins, 84, 2016
|
|
5VNB
 
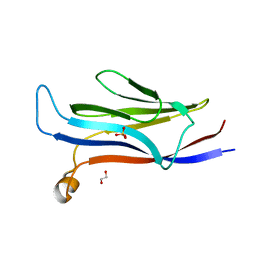 | | YEATS in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, H3K23acK27ac peptide, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
8GUJ
 
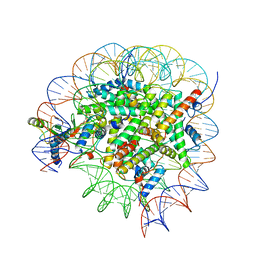 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
5DR6
 
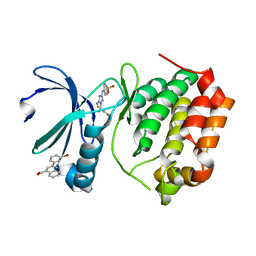 | | Aurora A Kinase in Complex with AA30 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)quinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
4AHU
 
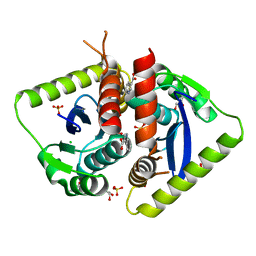 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOLE-3-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
8GUI
 
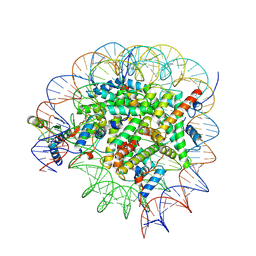 | | Bre1-nucleosome complex (Model I) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
5W58
 
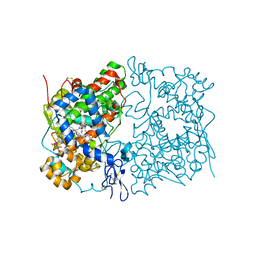 | | Crystal Complex of Cyclooxygenase-2: (S)-ARN-2508 (a dual COX and FAAH inhibitor) | | Descriptor: | (2S)-2-{2-fluoro-3'-[(hexylcarbamoyl)oxy][1,1'-biphenyl]-4-yl}propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Goodman, M.C, Banerjee, S, Piomelli, D, Marnett, L.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Dual cyclooxygenase-fatty acid amide hydrolase inhibitor exploits novel binding interactions in the cyclooxygenase active site.
J. Biol. Chem., 293, 2018
|
|
8GUK
 
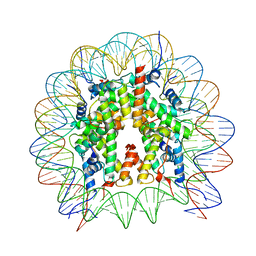 | | Human nucleosome core particle (free form) | | Descriptor: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
5C8V
 
 | |
7S8H
 
 | |
7S8G
 
 | |
5W66
 
 | | RNA polymerase I Initial Transcribing Complex State 3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
1MO7
 
 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
3TEX
 
 | |
8EKB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, deacylated P-site tRNAmet, and thermorubin at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Paranjpe, M.N, Polikanov, Y.S. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of translation inhibition by the ribosome-targeting antibiotic thermorubin.
Nucleic Acids Res., 51, 2023
|
|
4AHT
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
5VY5
 
 | | Rabbit muscle aldolase using 200keV | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Herzik Jr, M.A, Wu, M, Lander, G.C. | | Deposit date: | 2017-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Achieving better-than-3- angstrom resolution by single-particle cryo-EM at 200 keV.
Nat. Methods, 14, 2017
|
|
