8FNO
 
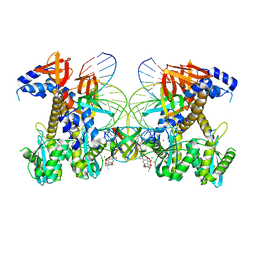 | | Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
6LMI
 
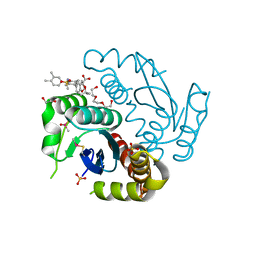 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-chromen-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase catalytic, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
1BIU
 
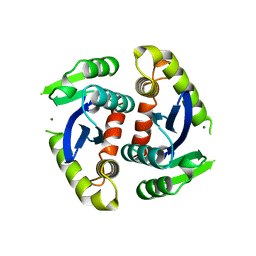 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6KDJ
 
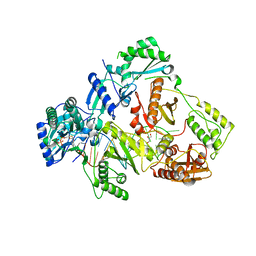 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
7T9H
 
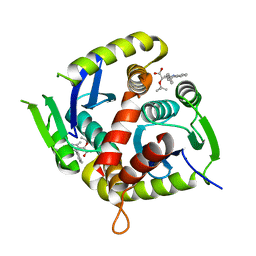 | | HIV Integrase in complex with Compound-15 | | Descriptor: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
6KDK
 
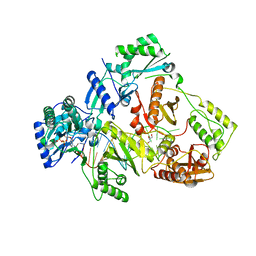 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDO
 
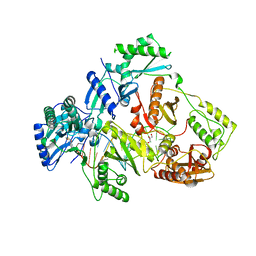 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
5XN0
 
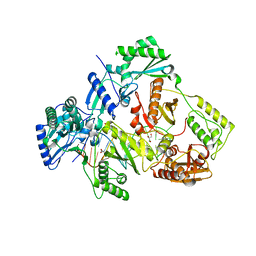 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
6IKA
 
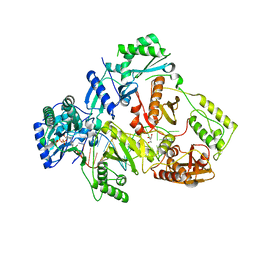 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
1A43
 
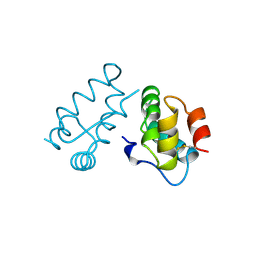 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAJ
 
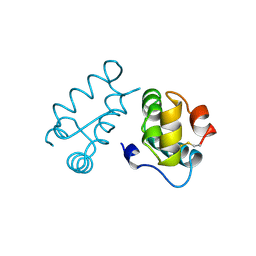 | | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ITG
 
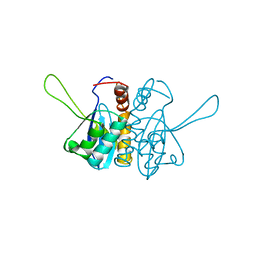 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
4ZHR
 
 | | Structure of HIV-1 RT Q151M mutant | | Descriptor: | RT p51 subunit, RT p66 subunit | | Authors: | Nakamura, A, Tamura, N, Yasutake, Y. | | Deposit date: | 2015-04-27 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the HIV-1 reverse transcriptase Q151M mutant: insights into the inhibitor resistance of HIV-1 reverse transcriptase and the structure of the nucleotide-binding pocket of Hepatitis B virus polymerase.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8X1Z
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
4GW6
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](tert-butoxy)ethanoic acid, ARSENIC, Gag-Pol polyprotein | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The A128T Resistance Mutation Reveals Aberrant Protein Multimerization as the Primary Mechanism of Action of Allosteric HIV-1 Integrase Inhibitors.
J.Biol.Chem., 288, 2013
|
|
5KGX
 
 | | HIV1 catalytic core domain in complex with an inhibitor (2~{S})-2-[3-(3,4-dihydro-2~{H}-chromen-6-yl)-1-methyl-indol-2-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-methyl-1H-indol-2-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Feng, L, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-10-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Indole-based allosteric inhibitors of HIV-1 integrase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7DBN
 
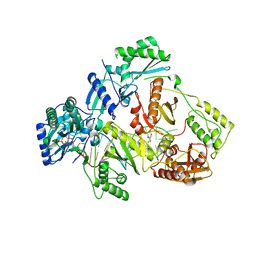 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
8X20
 
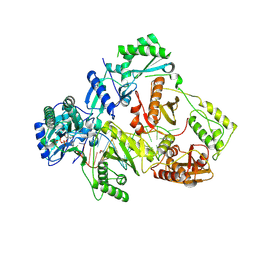 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8TOV
 
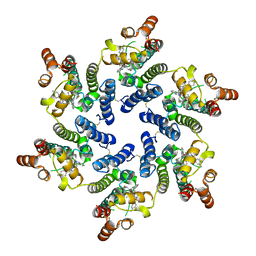 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-[(1R)-2-(3,5-difluorophenyl)-1-{3-[4-(morpholine-4-sulfonyl)phenyl]-4-oxo-3,4-dihydroquinazolin-2-yl}ethyl]acetamide, Matrix protein p17 | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-04 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
8FNN
 
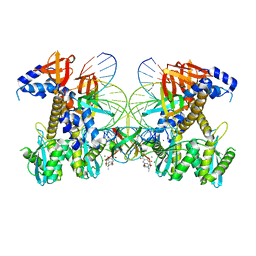 | | Structure of E138K/G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
3DS3
 
 | |
3H4E
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-19 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
6PUZ
 
 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUY
 
 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
