1J1W
 
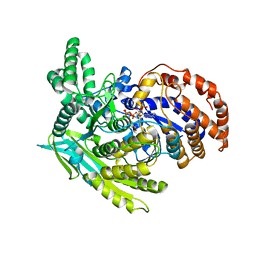 | | Crystal Structure Of The Monomeric Isocitrate Dehydrogenase In Complex With NADP+ | | Descriptor: | Isocitrate Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-12-19 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Monomeric Isocitrate Dehydrogenase in the Presence of NADP+
J.Biol.Chem., 278, 2003
|
|
1ITW
 
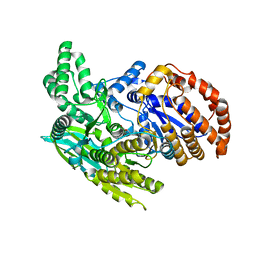 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-02-12 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
5C9I
 
 | |
4PB5
 
 | | D-threo-3-hydroxyaspartate dehydratase H351A mutant complexed with L-erythro-3-hydroxyaspartate | | Descriptor: | (3R)-3-hydroxy-L-aspartic acid, D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4PB3
 
 | | D-threo-3-hydroxyaspartate dehydratase H351A mutant | | Descriptor: | D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4PB4
 
 | | D-threo-3-hydroxyaspartate dehydratase H351A mutant complexed with 2-amino maleic acid | | Descriptor: | 2-amino maleic acid, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4YFB
 
 | |
4YF9
 
 | |
4YFA
 
 | |
2DTX
 
 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with D-mannose | | Descriptor: | Glucose 1-dehydrogenase related protein, SULFATE ION, beta-D-mannopyranose | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-18 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
2DTD
 
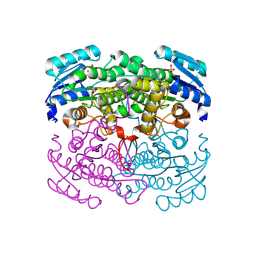 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in ligand-free form | | Descriptor: | Glucose 1-dehydrogenase related protein, SULFATE ION | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
2DTE
 
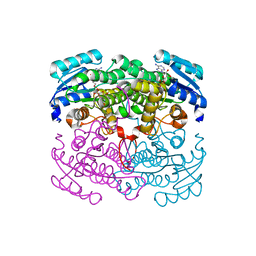 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase related protein | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
5GNM
 
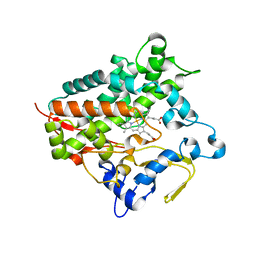 | | Cytochrome P450 Vdh (CYP107BR1) L348M mutant | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Yasutake, Y, Tamura, T. | | Deposit date: | 2016-07-22 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5GNL
 
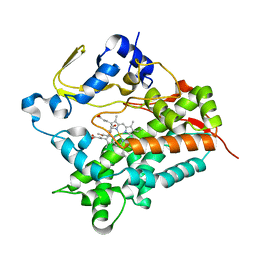 | | Cytochrome P450 Vdh (CYP107BR1) F106V mutant | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Yasutake, Y, Tamura, T. | | Deposit date: | 2016-07-22 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2Z36
 
 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
3A4H
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (orthorhombic crystal form) | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A51
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound 25-hydroxyvitamin D3 | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4G
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (trigonal crystal form) | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
7DBN
 
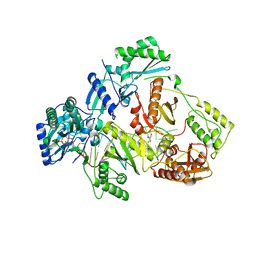 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
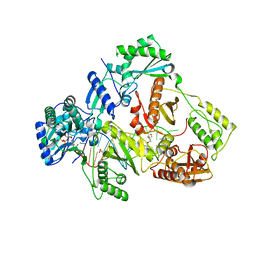 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7YM0
 
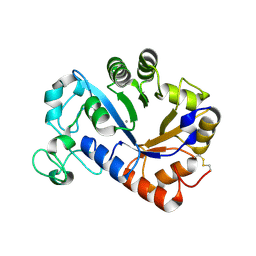 | | Lysoplasmalogen-specific phospholipase D (LyPls-PLD) with Ca2+ | | Descriptor: | CALCIUM ION, Lysoplasmalogenase | | Authors: | Yasutake, Y, Sakasegawa, S, Sugimori, D, Murayama, K. | | Deposit date: | 2022-07-27 | | Release date: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
5XN2
 
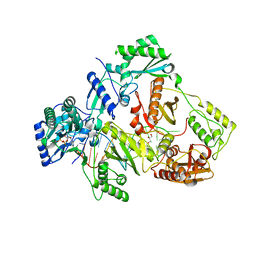 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
3WQE
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-allothreonine | | Descriptor: | D-allothreonine, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
