1U9L
 
 | | Structural basis for a NusA- protein N interaction | | Descriptor: | GOLD ION, Lambda N, Transcription elongation protein nusA | | Authors: | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-08-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1U9M
 
 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U9N
 
 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation opens therapeutic perspectives against tuberculosis and leprosy | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
1U9O
 
 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
1U9P
 
 | | Permuted single-chain Arc | | Descriptor: | pArc | | Authors: | Tabtiang, R.K, Cezairliyan, B.O, Grant, R.A, Cochrane, J.C, Sauer, R.T. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Consolidating critical binding determinants by noncyclic rearrangement of protein secondary structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1U9Q
 
 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
1U9R
 
 | |
1U9S
 
 | | Crystal structure of the specificity domain of Ribonuclease P of the A-type | | Descriptor: | BARIUM ION, RIBONUCLEASE P | | Authors: | Krasilnikov, A.S, Xiao, Y, Pan, T, Mondragon, A. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Basis for structural diversity in homologous RNAs.
Science, 306, 2004
|
|
1U9T
 
 | |
1U9U
 
 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U9V
 
 | |
1U9W
 
 | |
1U9X
 
 | |
1U9Y
 
 | | Crystal Structure of Phosphoribosyl Diphosphate Synthase from Methanocaldococcus jannaschii | | Descriptor: | Ribose-phosphate pyrophosphokinase | | Authors: | Kadziola, A, Johansson, E, Jepsen, C.H, McGuire, J, Larsen, S, Hove-Jensen, B. | | Deposit date: | 2004-08-11 | | Release date: | 2005-08-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
J.Mol.Biol., 354, 2005
|
|
1U9Z
 
 | | Crystal Structure of Phosphoribosyl Diphosphate Synthase Complexed with AMP and Ribose 5-Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, RIBOSE-5-PHOSPHATE, Ribose-phosphate pyrophosphokinase | | Authors: | Kadziola, A, Johansson, E, Jepsen, C.H, McGuire, J, Larsen, S, Hove-Jensen, B. | | Deposit date: | 2004-08-11 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
J.Mol.Biol., 354, 2005
|
|
1UA0
 
 | | Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
1UA1
 
 | | Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
1UA2
 
 | | Crystal Structure of Human CDK7 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein kinase 7 | | Authors: | Lolli, G, Lowe, E.D, Brown, N.R, Johnson, L.N. | | Deposit date: | 2004-08-11 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The Crystal Structure of Human CDK7 and Its Protein Recognition Properties
Structure, 12, 2004
|
|
1UA3
 
 | |
1UA4
 
 | | Crystal Structure of an ADP-dependent Glucokinase from Pyrococcus furiosus | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-dependent glucokinase, alpha-D-glucopyranose, ... | | Authors: | Ito, S, Jeong, J.J, Yoshioka, I, Koga, S, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-27 | | Release date: | 2004-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an ADP-dependent glucokinase from Pyrococcus furiosus: implications for a sugar-induced conformational change in ADP-dependent kinase
J.Mol.Biol., 331, 2003
|
|
1UA5
 
 | |
1UA6
 
 | | Crystal structure of HYHEL-10 FV MUTANT SFSF complexed with HEN EGG WHITE LYSOZYME complex | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-02-28 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.Biol.Chem., 278, 2003
|
|
1UA7
 
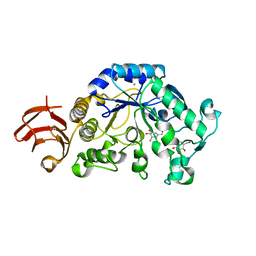 | | Crystal Structure Analysis of Alpha-Amylase from Bacillus Subtilis complexed with Acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, ... | | Authors: | Kagawa, M, Fujimoto, Z, Momma, M, Takase, K, Mizuno, H. | | Deposit date: | 2003-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bacillus subtilis alpha-amylase in complex with acarbose
J.BACTERIOL., 185, 2003
|
|
1UA8
 
 | | Crystal structure of the lipoprotein localization factor, LolA | | Descriptor: | Outer-membrane lipoproteins carrier protein | | Authors: | Takeda, K, Miyatake, H, Yokota, N, Matsuyama, S, Tokuda, H, Miki, K. | | Deposit date: | 2003-03-04 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of bacterial lipoprotein localization factors, LolA and LolB.
Embo J., 22, 2003
|
|
1UAA
 
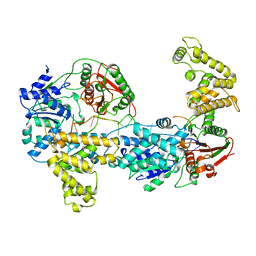 | | E. COLI REP HELICASE/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), PROTEIN (ATP-DEPENDENT DNA HELICASE REP.) | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP.
Cell(Cambridge,Mass.), 90, 1997
|
|
