1GKI
 
 | | Plasmid coupling protein TrwB in complex with ADP and Mg2+. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CONJUGAL TRANSFER PROTEIN TRWB, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
3PXF
 
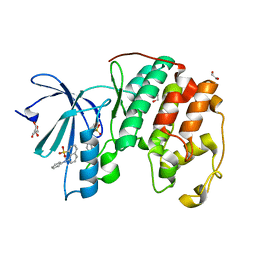 | | CDK2 in complex with two molecules of 8-anilino-1-naphthalene sulfonate | | Descriptor: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
1RPW
 
 | | Crystal Structure Of The Multidrug Binding Protein Qacr Bound To The Diamidine Hexamidine | | Descriptor: | 4,4'[1,6-HEXANEDIYLBIS(OXY)]BISBENZENECARBOXIMIDAMIDE, SULFATE ION, Transcriptional regulator qacR | | Authors: | Murray, D.S, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2003-12-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of QacR-diamidine complexes reveal additional multidrug-binding modes and a novel mechanism of drug charge neutralization.
J.Biol.Chem., 279, 2004
|
|
3QKX
 
 | |
3PXR
 
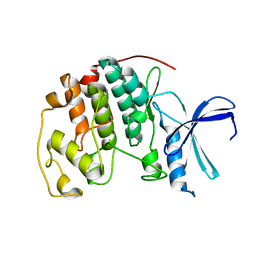 | | Apo CDK2 crystallized from Jeffamine | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PY0
 
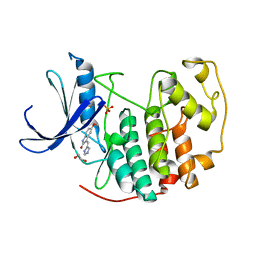 | | CDK2 in complex with inhibitor SU9516 | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, 1,2-ETHANEDIOL, Cell division protein kinase 2, ... | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PXY
 
 | | CDK2 in complex with inhibitor JWS648 | | Descriptor: | 2-(4,6-diamino-1,3,5-triazin-2-yl)-4-methoxyphenol, Cell division protein kinase 2, PHOSPHATE ION | | Authors: | Han, H, Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PXQ
 
 | | CDK2 in complex with 3 molecules of 8-anilino-1-naphthalene sulfonate | | Descriptor: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
2RA5
 
 | | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor | | Descriptor: | ISOPROPYL ALCOHOL, Putative transcriptional regulator, S,R MESO-TARTARIC ACID | | Authors: | Kim, Y, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
2BDE
 
 | | Crystal Structure of the cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) from Legionella pneumophila, Northeast Structural Genomics Target LgR1 | | Descriptor: | SULFATE ION, cytosolic IMP-GMP specific 5'-nucleotidase | | Authors: | Forouhar, F, Abashidze, M, Ho, C.K, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) from Legionella pneumophila, Northeast Structural Genomics Target LgR1
To be Published
|
|
4U2N
 
 | |
4U2M
 
 | |
6W2G
 
 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A in Monoclinic Unit Cell | | Descriptor: | 1,2-ETHANEDIOL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
6W2I
 
 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A | | Descriptor: | GLYCEROL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
4U8D
 
 | |
6VLI
 
 | | Crystal structure of transcriptional regulator from bacteriophage 186 | | Descriptor: | ACETATE ION, ACETIC ACID, Regulatory protein CII, ... | | Authors: | Truong, J.Q, Panjikar, S, Bruning, J.B, Shearwin, K.E. | | Deposit date: | 2020-01-24 | | Release date: | 2021-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.100021 Å) | | Cite: | Crystal structure of a transcriptional regulator from bacteriophage 186
To Be Published
|
|
6VMH
 
 | |
6VPE
 
 | |
2BZW
 
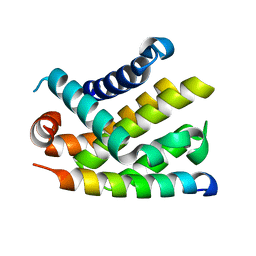 | | The crystal structure of BCL-XL in complex with full-length BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BCL2-ANTAGONIST OF CELL DEATH | | Authors: | Lee, K.-H, Han, W.-D, Kim, K.-J, Oh, B.-H. | | Deposit date: | 2005-08-24 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral Bcl-2 of Murine Gamma-Herpesvirus 68.
Plos Pathog., 4, 2008
|
|
2BSD
 
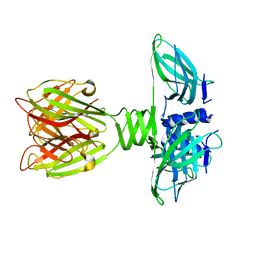 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein | | Descriptor: | RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, Dehaard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor-Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DA1
 
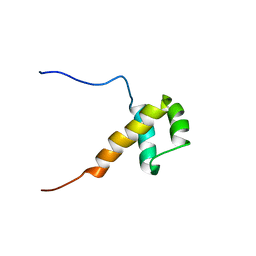 | | Solution structure of the first homeobox domain of AT-binding transcription factor 1 (ATBF1) | | Descriptor: | Alpha-fetoprotein enhancer binding protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first homeobox domain of AT-binding transcription factor 1 (ATBF1)
To be Published
|
|
4TWJ
 
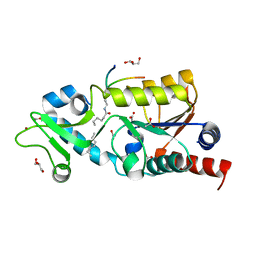 | | The structure of Sir2Af2 bound to a myristoylated histone peptide | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4 peptide, ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
4TYL
 
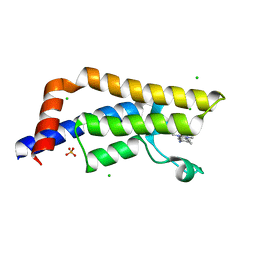 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 5-amino-1,3,6-trimethyl-1,3-dihydro-2H-benzimidazol-2-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
4TZ2
 
 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(5-phenyl-4H-1,2,4-triazol-3-yl)aniline, ATPase family AAA domain-containing protein 2, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
2DA3
 
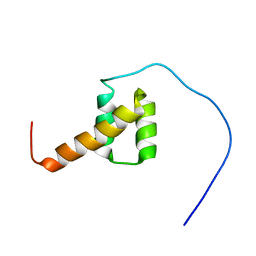 | | Solution structure of the third homeobox domain of AT-binding transcription factor 1 (ATBF1) | | Descriptor: | Alpha-fetoprotein enhancer binding protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third homeobox domain of AT-binding transcription factor 1 (ATBF1)
To be Published
|
|
