6W9O
 
 | |
1E0Q
 
 | | Mutant Peptide from the first N-terminal 17 amino-acid of Ubiquitin | | Descriptor: | POLYUBIQUITIN-B | | Authors: | Zerella, R, Chen, P.Y, Evans, P.A, Raine, A, Williams, D.H. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a Mutant Peptide Derived from Ubiquitin: Implications for Protein Folding.
Protein Sci., 9, 2000
|
|
6W9R
 
 | |
1DJA
 
 | |
4NVN
 
 | | Predicting protein conformational response in prospective ligand discovery | | Descriptor: | 2,3-dihydrobenzo[h][1,6]naphthyridin-4(1H)-one, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
4NVE
 
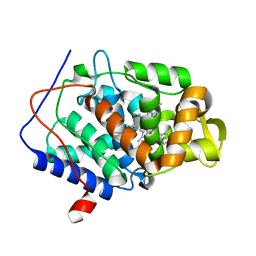 | |
4NVO
 
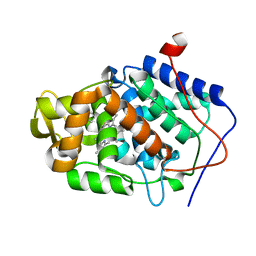 | |
2I4C
 
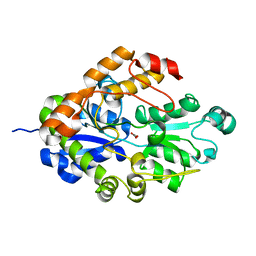 | | Crystal structure of Bicarbonate Transport Protein CmpA from Synechocystis sp. PCC 6803 in complex with bicarbonate and calcium | | Descriptor: | BICARBONATE ION, Bicarbonate transporter, CALCIUM ION | | Authors: | Koropatkin, N.M, Smith, T.J, Pakrasi, H.B. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of a Cyanobacterial Bicarbonate Transport Protein, CmpA.
J.Biol.Chem., 282, 2007
|
|
6B2X
 
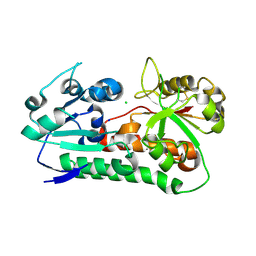 | | Apo YiuA Crystal Form 1 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4NBM
 
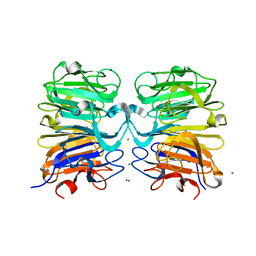 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
3A6N
 
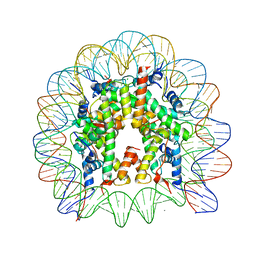 | | The nucleosome containing a testis-specific histone variant, human H3T | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | Deposit date: | 2009-09-04 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4N84
 
 | | Crystal structure of 14-3-3zeta in complex with a 12-carbon-linker cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2Y4V
 
 | |
2BIY
 
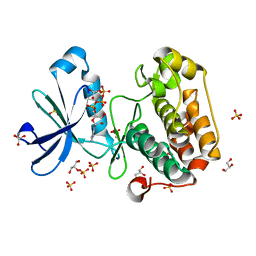 | | Structure of PDK1-S241A mutant kinase domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2005-01-26 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of T-loop phosphorylation in PDK1 activation, stability, and substrate binding.
J. Biol. Chem., 280, 2005
|
|
7E42
 
 | |
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
7S55
 
 | |
4P2A
 
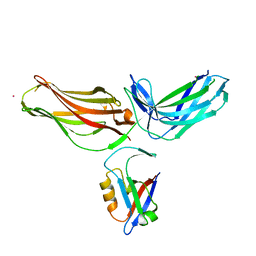 | | Structure of mouse VPS26A bound to rat SNX27 PDZ domain | | Descriptor: | MERCURY (II) ION, Sorting nexin-27, Vacuolar protein sorting-associated protein 26A | | Authors: | Clairfeuille, T, Gallon, M, Mas, C, Ghai, R, Teasdale, R, Cullen, P, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PAN
 
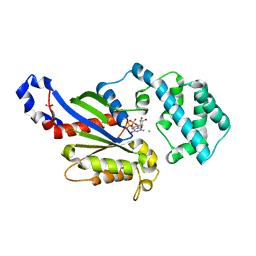 | | A conserved phenylalanine as relay between the 5 helix and the GDP binding region of heterotrimeric G protein | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Conserved Phenylalanine as a Relay between the alpha 5 Helix and the GDP Binding Region of Heterotrimeric Gi Protein alpha Subunit.
J.Biol.Chem., 289, 2014
|
|
2J4Z
 
 | | Structure of Aurora-2 in complex with PHA-680626 | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)-N-[5-(2-THIENYLACETYL)-1,5-DIHYDROPYRROLO[3,4-C]PYRAZOL-3-YL]BENZAMIDE, ARSENIC, SERINE THREONINE-PROTEIN KINASE 6 | | Authors: | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | Deposit date: | 2006-09-08 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile.
J. Med. Chem., 49, 2006
|
|
6B2Y
 
 | | Apo YiuA Crystal Form 2 | | Descriptor: | SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4P66
 
 | | Electrostatics of Active Site Microenvironments of E. coli DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
7SN3
 
 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
1DDS
 
 | |
1DDR
 
 | |
